

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| SPAC17A5.04c | SP | 0.000169 | 0.999827 | 0.000004 | CS pos: 17-18. SYA-EI. Pr: 0.8072 |
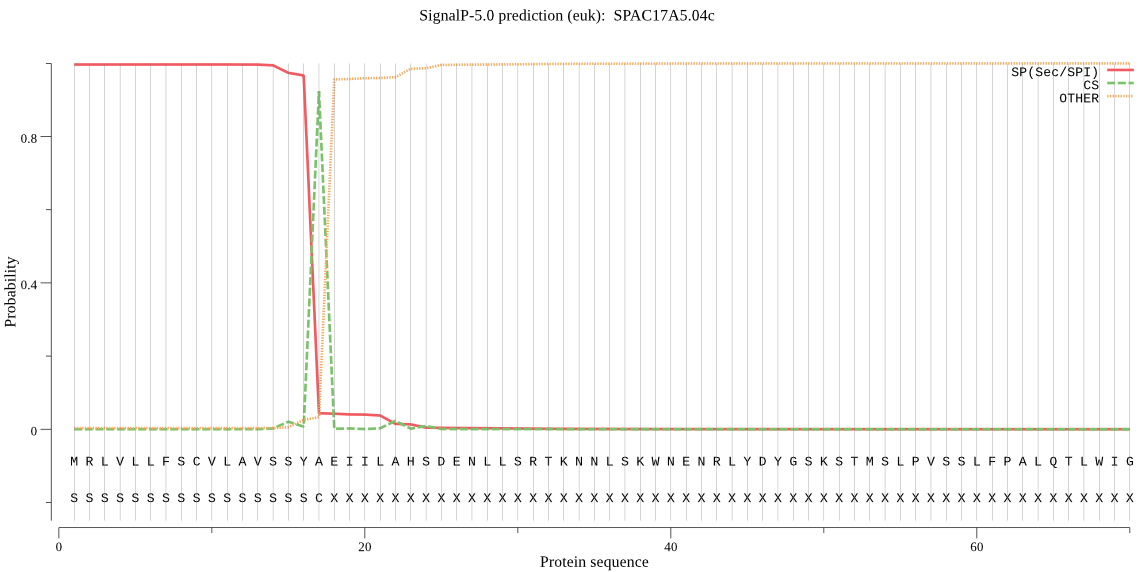
MRLVLLFSCVLAVSSYAEIILAHSDENLLSRTKNNLSKWNENRLYDYGSKSTMSLPVSSL FPALQTLWIGVVADCSYVTHFTSRMEAKKHIFQEFEGVSTLYEDSFNINVQIHSLILPSA HDCSANVVDRPEISMSPRISIEEKLEIFSKWKYESPGNNVFEAISPHERESFPSEPQVSV LFTSSVKRSPHGVSWFATICSETHIENEWHVGPLSVVSAYPNDRLVVAHEIGHILGLIHD CNKKSCGDHSEACCPLSSSLCDAQELYIMNPSNSYTYANLRFSDCSILQLHSLVEKKYVS LSCLSKPSEKSVLRLGTCGNGIVEDGEECDCGEDCENNPCCDGKTCKLTKGSLCDDQQDA CCYQCHFKNAGTLCRQSTNPCDKPEFCTGISSKCPVDENWDDGRICQDSLGMGSCASGVC TSASRQCKKLTNFSSLSCHSDSCKVSCQNEDGTCFISAKDYIDGTRCRGGLCYNGVCVPI EGSSASWSKQPSLFCASGTMLISLAVIAWFFW
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| SPAC17A5.04c | 24 S | ILAHSDENL | 0.991 | unsp | SPAC17A5.04c | 140 S | SPRISIEEK | 0.997 | unsp |