

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
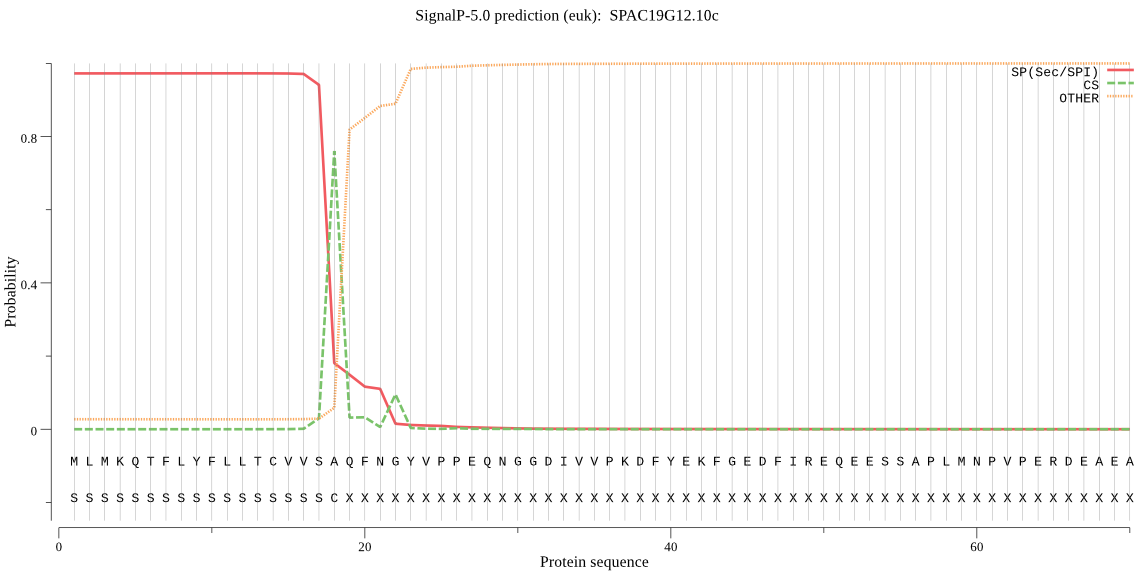
| SPAC19G12.10c | SP | 0.009086 | 0.990814 | 0.000100 | CS pos: 18-19. VSA-QF. Pr: 0.7748 |
MLMKQTFLYFLLTCVVSAQFNGYVPPEQNGGDIVVPKDFYEKFGEDFIREQEESSAPLMN PVPERDEAEAPHHPKGHHEFNDDFEDDTALEHPGFKDKLDSFLQPARDFLHTVSDRLDNI FDDDEDEHVREKRPHDSADEDAPRRKHGKCKGKGKHHKGKHAKGKGKKSHPKPEDDSVFF DDERPKHHEFDDEDREFPAHHEPGEHMPPPPMHHKPGEHMPPPPMHHEPGEHMPPPPMHH EPGEHMPPPPMHHEPGEHMPPPPMHHEPGEHMPPPPMHHEPGEHMPPPPMHHEPGEHMPP PPMHHEPGEHMPPPPMHHEPGEHMPPPPMHHEPGEHMPPPPFKHHELEEHEGPEHHRGPE DKEHHKGPKDKEHHKGPKDKEHHKGPKDKEHHKGPKDKEHHKGPKDKEHHKGPKDKEHHQ GPKEKHNERPEQNMQSSHELLVIEAFADLINSVPVEEIAEEFSRFLDTLGIEYYGNIPVH IQENAPKDSSIPPLFEFDDDLELSDLTPEQFAYLEMLKAEGIDPMTAFRDQSHPAKPSNA QPADSSRPYAVFSQEENGEHVNLKAFPDHTLRVKDSKPESLGIDTVKQYTGYLDVEDDRH LFFWFFESRNDPENDPVVLWLNGGPGCSSLTGLFMELGPSSINIETLKPEYNPHSWNSNA SVIFLDQPINTGFSNGDDSVLDTVTAGKDVYAFLNLFFAKFPQYAHLDFHIAGESYAGHY IPQFAKEIMEHNQGANFFVASGYEMEKQYINLKSVLIGNGLTDPLVQYYFYGKMACESPY GPIMSQEECDRITGAYDTCAKLITGCYQTGFTPVCIGASLYCNNAMIGPFTKTGLNIYDI REECRDQEHLCYPETGAIESYLNQEFVQEALGVEYDYKGCNTEVNIGFLFKGDWMRKTFR DDVTAILEAGLPVLIYAGDADYICNYMGNEAWTDALEWAGQREFYEAELKPWSPNGKEAG RGKSFKNFGYLRLYEAGHMVPFNQPEASLEMLNSWIDGSLFA
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SPAC19G12.10c | 679 S | NGDDSVLDT | 0.997 | unsp | SPAC19G12.10c | 679 S | NGDDSVLDT | 0.997 | unsp | SPAC19G12.10c | 679 S | NGDDSVLDT | 0.997 | unsp | SPAC19G12.10c | 898 T | WMRKTFRDD | 0.991 | unsp | SPAC19G12.10c | 553 S | YAVFSQEEN | 0.993 | unsp | SPAC19G12.10c | 674 S | NTGFSNGDD | 0.992 | unsp |