

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
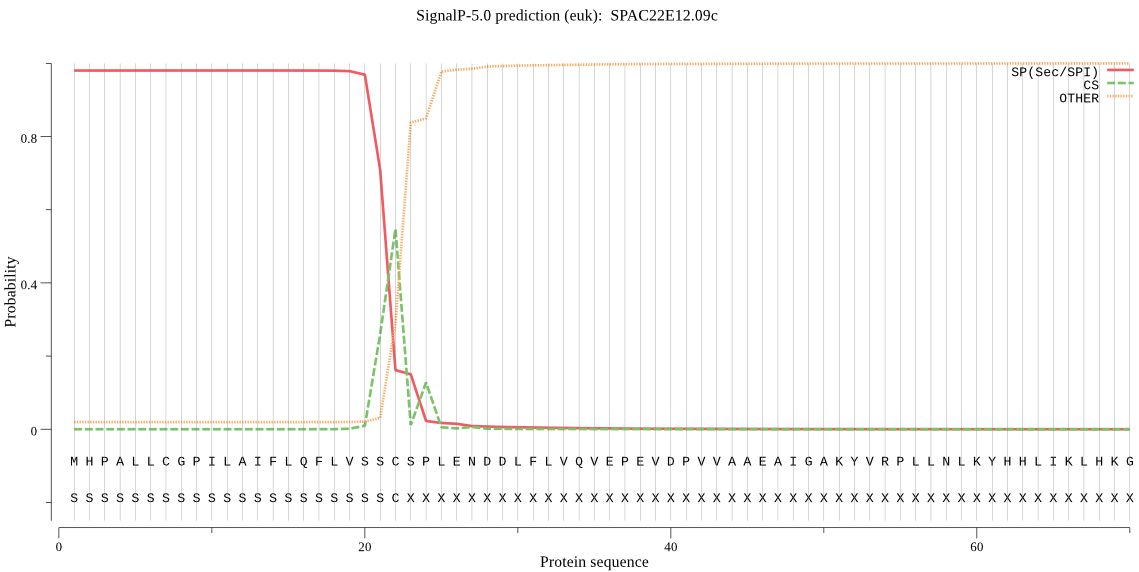
| SPAC22E12.09c | SP | 0.015335 | 0.984608 | 0.000057 | CS pos: 22-23. SSC-SP. Pr: 0.6461 |
MHPALLCGPILAIFLQFLVSSCSPLENDDLFLVQVEPEVDPVVAAEAIGAKYVRPLLNLK YHHLIKLHKGSDDSVQSSIRKRGIDAGILELERQTPRWRYKRDASESDELLNEFSNHFGI SDPLFYGQWHIFNSNNPGHDLNLREVWDAGYFGENVTVAFVDDGIDFKHPDLQAAYTSLG SWDFNDNIADPLPKLSDDQHGTRCAGEVAAAWNDVCGVGIAPRAKVAGLRILSAPITDAV ESEALNYGFQTNHIYSCSWGPADDGRAMDAPNTATRRALMNGVLNGRNGLGSIFVFASGN GGHYHDNCNFDGYTNSIFSATIGAVDAEHKIPFYSEVCAAQLVSAYSSGSHLSILTTNPE GTCTRSHGGTSAAAPLASAVYALALSIRPDLSWRDIQHITVYSASPFDSPSQNAEWQKTP AGFQFSHHFGFGKLDASKFVEVAKDWQVVNPQTWLIAPEINVNKSFGSVNNETITEMVSE FTVTKDMIEKSNFKRLEHVTVRVCIPFNRRGALEILLESPSGIRSILASERPYDENSKGF LDWTFMTVQHWAEPPEGVWKLLVNDRSGGKHEGTFENWQLALWGESENPSNTAPLPYDTL ELPKEMVLGIYSEPNSDLTNSSTLLSPTSTSFTSYTVSATATPTSTSHIPIPTVLPPTQP VLEPSYREIVAFITFFLLFAFIFVAVIWTWISAFWKAKAPPPLSQQEIA
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SPAC22E12.09c | 392 S | RPDLSWRDI | 0.997 | unsp | SPAC22E12.09c | 392 S | RPDLSWRDI | 0.997 | unsp | SPAC22E12.09c | 392 S | RPDLSWRDI | 0.997 | unsp | SPAC22E12.09c | 405 S | VYSASPFDS | 0.995 | unsp | SPAC22E12.09c | 665 S | VLEPSYREI | 0.991 | unsp | SPAC22E12.09c | 95 T | LERQTPRWR | 0.995 | unsp | SPAC22E12.09c | 105 S | KRDASESDE | 0.997 | unsp |