

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| SPAC22F8.06 | OTHER | 0.999984 | 0.000011 | 0.000005 |
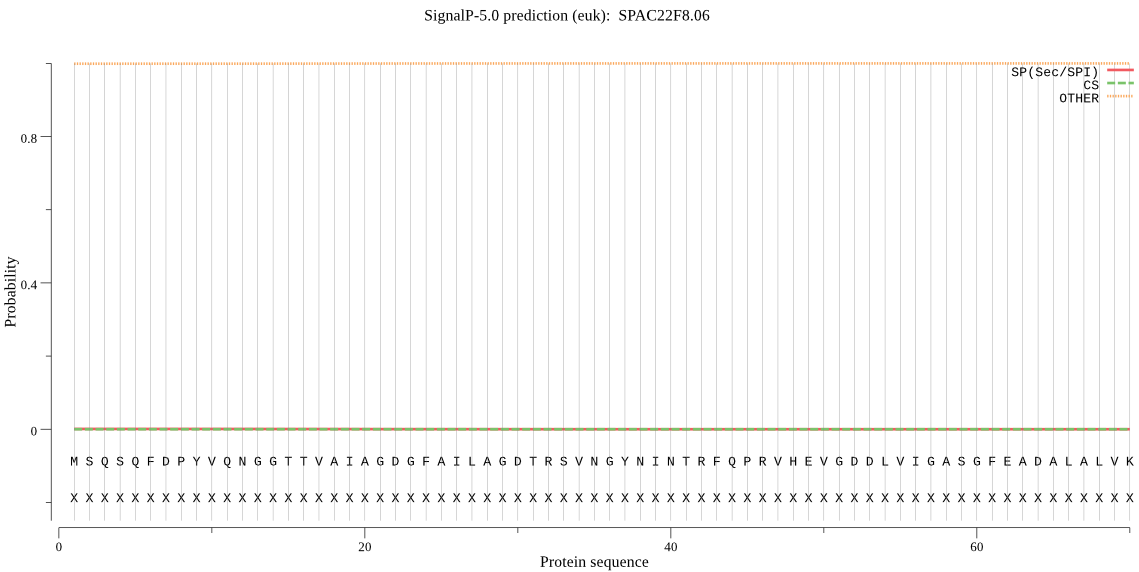
MSQSQFDPYVQNGGTTVAIAGDGFAILAGDTRSVNGYNINTRFQPRVHEVGDDLVIGASG FEADALALVKRIQQRIDLYHDNHERKMSAQSCACMVRTLLYGKRFFPYYVYTTVAGIDKE GKGEIYSFDPVGSYEREWCRAGGSAANFITPFLDNQVNLHNQYVPGSHGKERKPRRLLKL EEAMKITTDAFTSAGERHIEVGDSVLVKIITKEGVETRIIPLKKD