

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| SPAC23D3.07 | OTHER | 0.998828 | 0.000457 | 0.000715 |
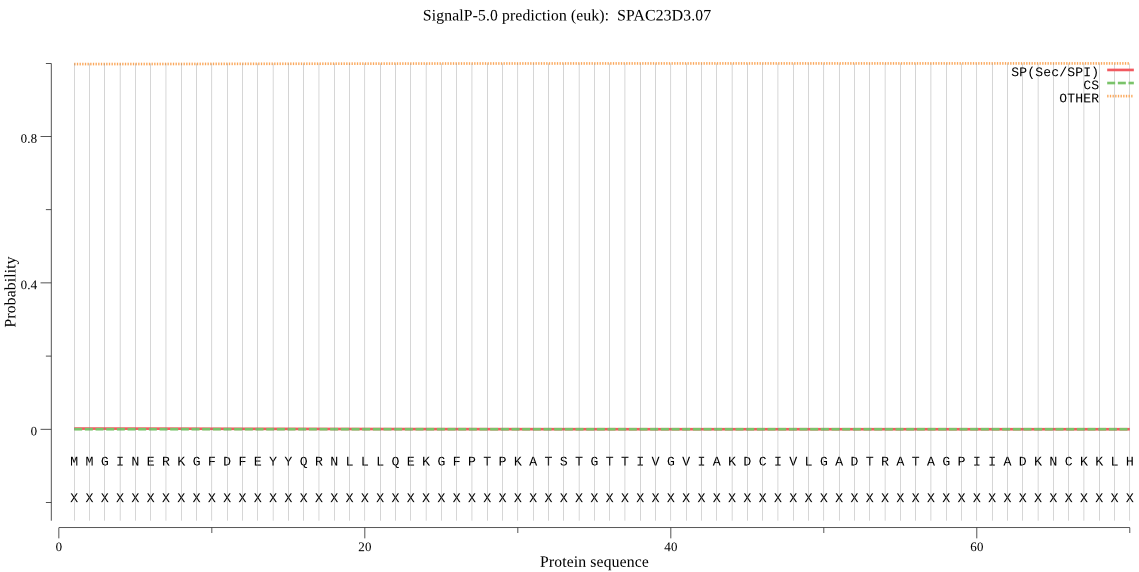
MMGINERKGFDFEYYQRNLLLQEKGFPTPKATSTGTTIVGVIAKDCIVLGADTRATAGPI IADKNCKKLHLISPNIWCAGAGTAADTEFVTSMISSNIELHSLYTNRKPRVVTALTMLKQ HLFRYQGHIGAYLVLGGYDCKGPHLFTIAAHGSSDKLPYVALGSGSLAAISVLETKYQPD LERHEAMELVKEAIEAGIFNDLGSGSNCDLVVIDEEKATPYRGYSKPNERATKQSKYTYD RGTTAVLKEDIYKFVTVQDLDEMQVDV