

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
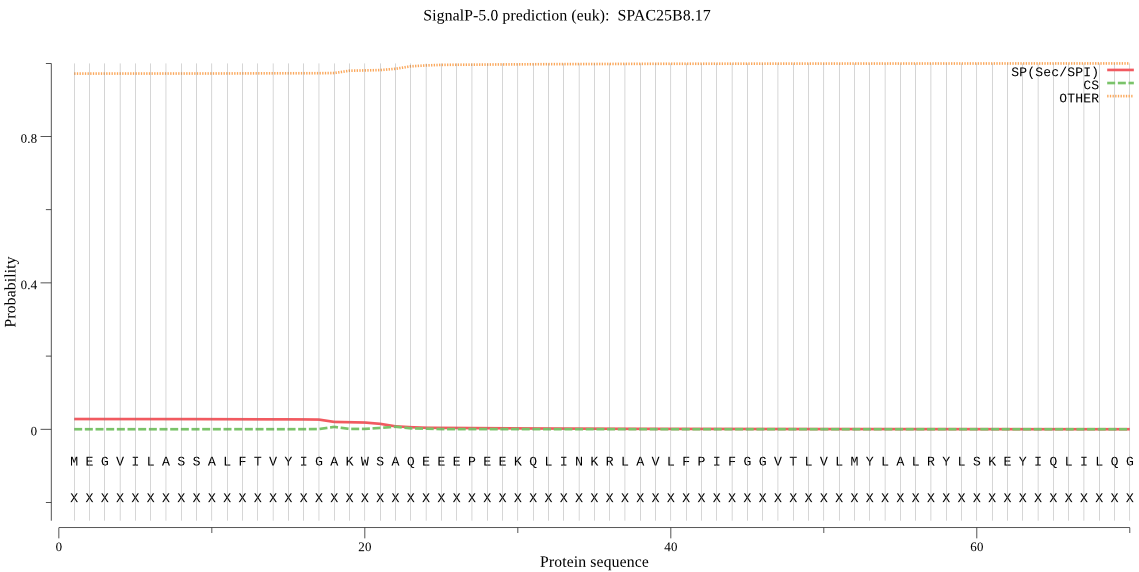
| SPAC25B8.17 | OTHER | 0.983095 | 0.016641 | 0.000263 |
MEGVILASSALFTVYIGAKWSAQEEEPEEKQLINKRLAVLFPIFGGVTLVLMYLALRYLS KEYIQLILQGYASLASIICFVRSFNPKTTFGKITATMSSIAIALFYFKTKHWMASNILAW ALAANSISIMRIDSYNTGALLLGALFFYDIYFVFGTEVMVTVATGIDIPAKYVLPQFKNP TRLSMLGLGDIVMPGLMLALMYRFDLHYYINSTSQPKKHSTYFRNTFIAYGLGLGVTNFA LYYFKAAQPALLYLSPACIVAPLLTAWYRDELKTLFSFRSETEDETDEQDKCKST
| ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|
| SPAC25B8.17 | T | 295 | 0.605 | 0.022 |
| ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|
| SPAC25B8.17 | T | 295 | 0.605 | 0.022 |