

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
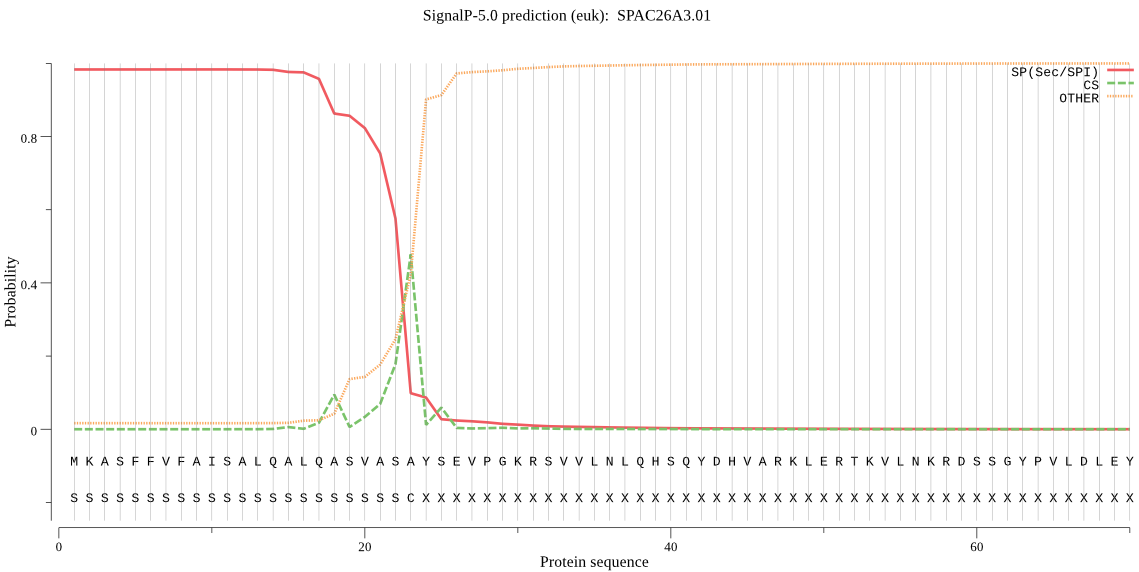
| SPAC26A3.01 | SP | 0.000776 | 0.998958 | 0.000266 | CS pos: 23-24. ASA-YS. Pr: 0.3736 |
MKASFFVFAISALQALQASVASAYSEVPGKRSVVLNLQHSQYDHVARKLERTKVLNKRDS SGYPVLDLEYTDAGGYFANLTLGSNERVYSLTLDTGSPYTWVTAKNITALSASEIWSDTD GVDAGRSTSDIRTNACTNYTCFDYSSTTARRTNSSTIGFLASYGDNTTVLGYNMVDNAYF AGLTLPGFEFGLATREYDSSQISVTPGIIGLSVAMTITGISSDDKVVAFTPPTIVDQLVS ANVIDTPAFGIYLNEDVGELIFGGYDKAKINGSVHWVNISSSDDSTFYSVNLESITVTNS TSSNNVQSSKRSSKDIEVNTTVTLDTGTVYIYLPEDTVESIADQYQGIVSEYGYVVIYCD SFSDSDYISFNFGSDADFHVSVNDLVIYRQESTSGDICYLALFEGDTSSYLLGQYFLQYV YSIYDWDAQKIGLAALNSNATSTANHQILNINSALRSVTSGQSVSATPTVSMSIAATSFG SSLVLTASASPSSTSVDGSSSSDSSEASGAASVGVSISAIVLCASTLISLLFA
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SPAC26A3.01 | 312 S | SSKRSSKDI | 0.998 | unsp | SPAC26A3.01 | 312 S | SSKRSSKDI | 0.998 | unsp | SPAC26A3.01 | 312 S | SSKRSSKDI | 0.998 | unsp | SPAC26A3.01 | 313 S | SKRSSKDIE | 0.994 | unsp | SPAC26A3.01 | 392 S | YRQESTSGD | 0.995 | unsp | SPAC26A3.01 | 500 S | VDGSSSSDS | 0.996 | unsp | SPAC26A3.01 | 504 S | SSSDSSEAS | 0.996 | unsp | SPAC26A3.01 | 40 S | NLQHSQYDH | 0.996 | unsp | SPAC26A3.01 | 127 S | DAGRSTSDI | 0.992 | unsp |