

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| SPAC323.02c | OTHER | 0.999765 | 0.000089 | 0.000147 |
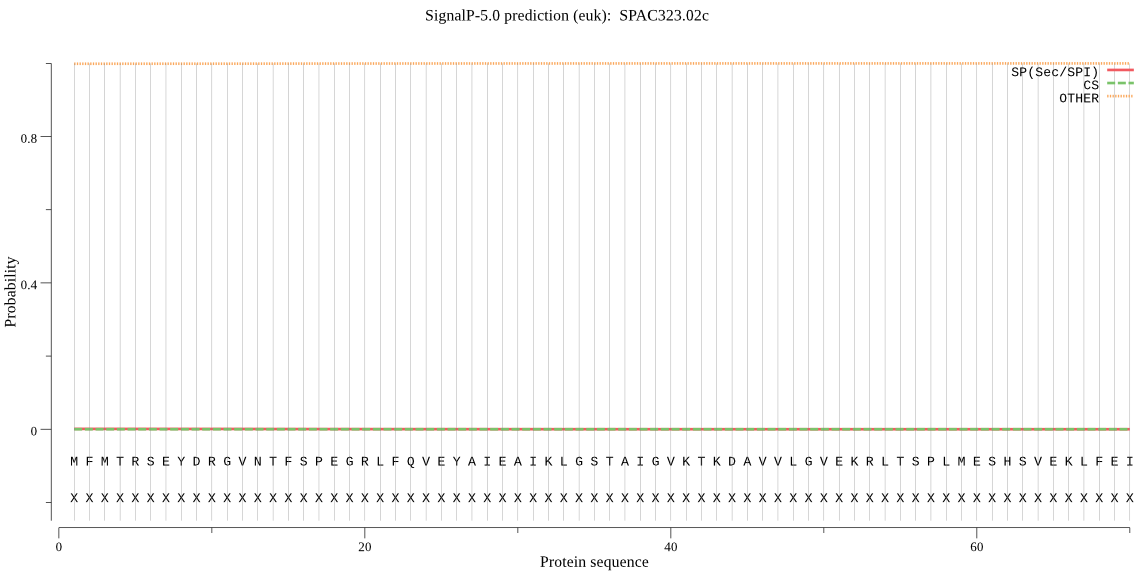
MFMTRSEYDRGVNTFSPEGRLFQVEYAIEAIKLGSTAIGVKTKDAVVLGVEKRLTSPLME SHSVEKLFEIDSHIGCAISGLTADARTIIEHARVQTQNHRFTYDEPQGIESTTQSICDLA LRFGEGEDGEERIMSRPFGVALLIAGIDEHGPQLYHSEPSGTYFRYEAKAIGSGSEPAKS ELVKEFHKDMTLEEAEVLILKVLRQVMEEKLDSKNVQLAKVTAEGGFHIYNDEEMADAVA REQQRMD