

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| SPAC4A8.04 | SP | 0.003688 | 0.995486 | 0.000826 | CS pos: 24-25. ACA-AP. Pr: 0.7236 |
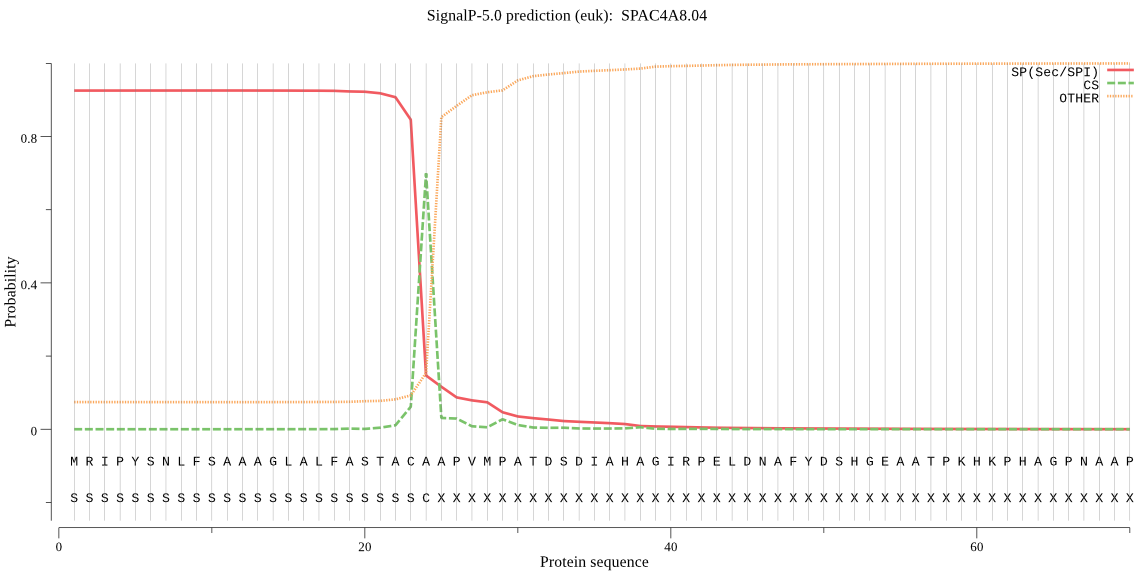
MRIPYSNLFSAAAGLALFASTACAAPVMPATDSDIAHAGIRPELDNAFYDSHGEAATPKH KPHAGPNAAPLLSASNADTTGLDSHYIIVLQPDLSEQEFQAHTNWVSEMHQMDIASQEDE YYDTSDSNYMFGLKHVYDFGEDSFKGYSGQFSSNIVEQIRLHPHVIAVERDQVVSIKKLE TQSGAPWGLARISHKSVKYDDIGKYVYDSSAGDNITAYVVDTGVSIHHVEFEGRASWGAT IPSGDVDEDNNGHGTHVAGTIASRAYGVAKKAEIVAVKVLRSSGSGTMADVIAGVEWTVR HHKSSGKKTSVGNMSLGGGNSFVLDMAVDSAVTNGVIYAVAAGNEYDDACYSSPAASKKA ITVGASTINDQMAYFSNYGSCVDIFAPGLNILSTWIGSNTSTNTISGTSMATPHVAGLSA YYLGLHPAASASEVKDAIIKMGIHDVLLSIPVGSSTINLLAFNGAQE
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SPAC4A8.04 | 236 S | EGRASWGAT | 0.995 | unsp | SPAC4A8.04 | 236 S | EGRASWGAT | 0.995 | unsp | SPAC4A8.04 | 236 S | EGRASWGAT | 0.995 | unsp | SPAC4A8.04 | 283 S | VLRSSGSGT | 0.995 | unsp | SPAC4A8.04 | 305 S | HHKSSGKKT | 0.996 | unsp | SPAC4A8.04 | 357 S | SPAASKKAI | 0.992 | unsp | SPAC4A8.04 | 193 S | LARISHKSV | 0.992 | unsp | SPAC4A8.04 | 196 S | ISHKSVKYD | 0.994 | unsp |