

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
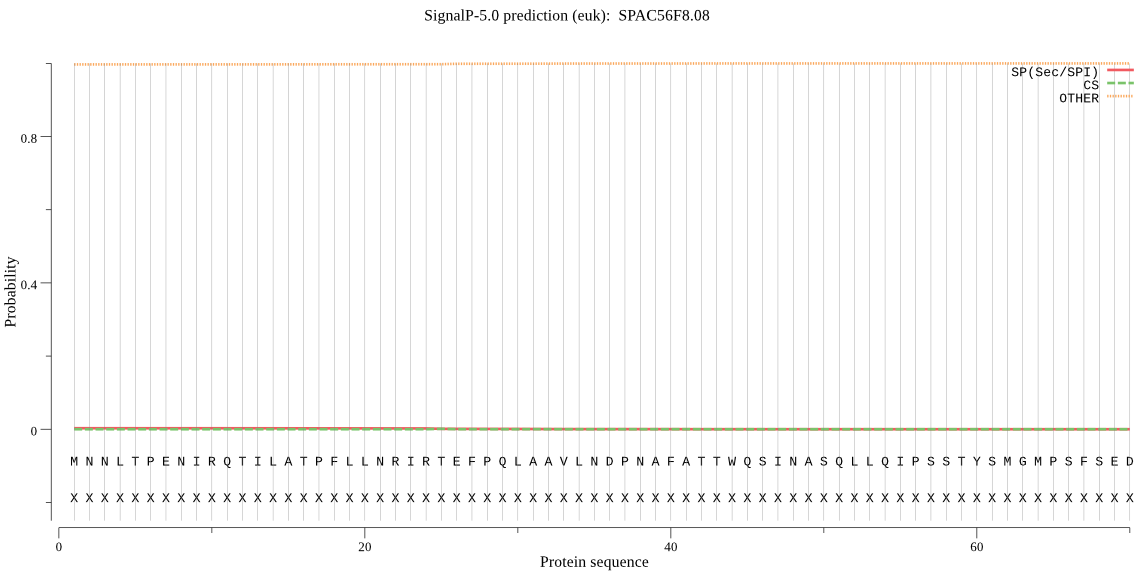
| SPAC56F8.08 | OTHER | 0.954295 | 0.015686 | 0.030019 |
MNNLTPENIRQTILATPFLLNRIRTEFPQLAAVLNDPNAFATTWQSINASQLLQIPSSTY SMGMPSFSEDDLFDVEVQRRIEEQIRQNAVTENMQSAIENHPEVFGQVYMLFVNVEINGH KVKAFVDSGAQATILSADCAEKCGLTRLLDTRFQGVAKGVGMAKILGCVHSAPLKIGDLY LPCRFTVIEGRDVDMLLGLDMLRRYQACIDLENNVLRIHGKEIPFLGESEIPKLLANVEP SANAHGLGIEPASKASASSPNPQSGTRLGTKESVAPNNEGSSNPPSLVNPPTDPGLNSKI AQLVSMGFDPLEAAQALDAANGDLDVAASFLL
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SPAC56F8.08 | T | 270 | 0.663 | 0.050 | SPAC56F8.08 | T | 266 | 0.637 | 0.024 | SPAC56F8.08 | S | 273 | 0.532 | 0.049 | SPAC56F8.08 | S | 264 | 0.502 | 0.021 |
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SPAC56F8.08 | T | 270 | 0.663 | 0.050 | SPAC56F8.08 | T | 266 | 0.637 | 0.024 | SPAC56F8.08 | S | 273 | 0.532 | 0.049 | SPAC56F8.08 | S | 264 | 0.502 | 0.021 |
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| SPAC56F8.08 | 68 S | MPSFSEDDL | 0.992 | unsp |