

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
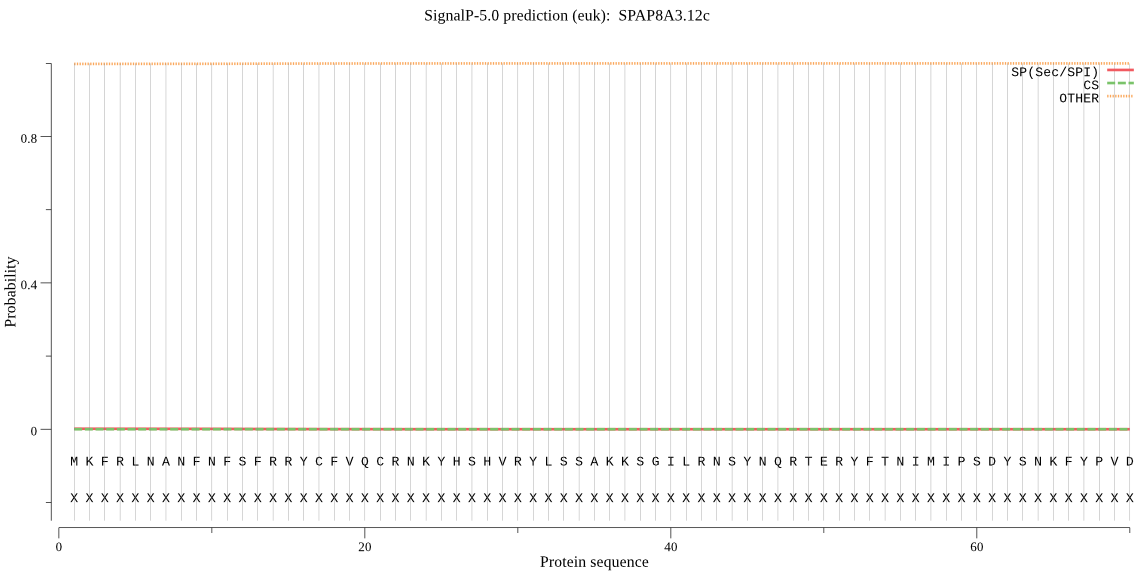
| SPAP8A3.12c | mTP | 0.315122 | 0.000033 | 0.684846 | CS pos: 31-32. VRY-LS. Pr: 0.4698 |
MKFRLNANFNFSFRRYCFVQCRNKYHSHVRYLSSAKKSGILRNSYNQRTERYFTNIMIPS DYSNKFYPVDGVVPKHETQAYEFLKKFPEYDGRGVTVGILDTGVDPGAPGLSVTTTGLPK FKNIVDCTGAGDVDTSVEVAAADSNDYLTITGRSGRTLKLSKEWKNPSKKWKVGCKLAYE FFPKDLRKRLQKLETEDMNKSNRKLLQDATDEYAKFKDKFPEAPLDKDNLQTQKELEARI ECLKQLAEKFDNPGPLYDVVVFHDGEHWRVVIDSDQTGDIYLHKPLADFNVAQEWSTFGS LDLLSYGVHVYDNGNITSIVAVSGTHGTHVAGIIGANHPETPELNGAAPGCQLVSLMIGD GRLDSLETSHAFSRACSEIIKNEVDIINISFGEDAGIPNKGRVIELLRDELAGKRNVVIV SSAGNNGPAYTTVGAPGGTTFDVISVGAYVTSGMMQAQYNLLSTVHDTPYTWCSRGPTLD GDTGVSIYAPGGAITSVPPYSLQNSQLMNGTSMSSPSACGGISLILSALKAQKKPYTAAA IKKAVMYTSKDLRDDFNTGMLQVDNAYEYLAQSDFQYTGARSFTINGNIGNSKRGVYLRN PTEVCSPSRHMFNVAPKFEDGEEYEKSHFEVQLSLATTQPWIQAPEYVMMAGTGRGIPVR VDPTALAPGHHFGKVLAYDASNESRRCVFEIPVTVMKPSSISNTCFSLRDVSFEPTLIKR HFLVPPKGATYVEIRVKATSELESTNMLWISVNQTIPQTKLNEASTELIMPVTQNEVTTK LVSIDDSYTLELCMAQWWSSLEPMVLDIDVNFHGIKVVNGKEINLISSQGLKRVDCASIR RENFKPDITLKDYVDSFKPTNTVIKPLGDRDIMPDGQQLFELMATYSVEISEKTELKADF AVPHNMYDNGFNGLFFMVFDSQKQRVHYGDMYTSSHTLEKGEYLYKFQLLSVDPSTLERF RNVTLRLTKKLKKPITLPLYADHIDFCDNKTYERENIDAGVVESFVVGTNIEGEQYASEL KENSLLTGELKFGDCEKGTVPVTLVLPPKISTKEDTKLGEKCANIVQLQVDLLSKLADQE KEKHLKYLQSSYKNSLEVQLAKLDIVKETNERLSTADSILSLIDTEALSRYYSCQQKVED TIPRDVVLEKKMALQRDAFIRALVVKCETFSTQGHKDKDNYFQNYQLLLNWLENSDPRVW QIKKDYYKSQNQYGLALKALLELLKENGNSGKMDVAKLLSEEKELLVNLGWNYWHDIVFV ETVKRVPPYSYALF
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SPAP8A3.12c | 740 S | VKATSELES | 0.996 | unsp | SPAP8A3.12c | 740 S | VKATSELES | 0.996 | unsp | SPAP8A3.12c | 740 S | VKATSELES | 0.996 | unsp | SPAP8A3.12c | 783 S | TKLVSIDDS | 0.996 | unsp | SPAP8A3.12c | 1091 S | YLQSSYKNS | 0.995 | unsp | SPAP8A3.12c | 1114 S | NERLSTADS | 0.998 | unsp | SPAP8A3.12c | 1121 S | DSILSLIDT | 0.994 | unsp | SPAP8A3.12c | 27 S | NKYHSHVRY | 0.993 | unsp | SPAP8A3.12c | 712 S | LRDVSFEPT | 0.994 | unsp |