

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
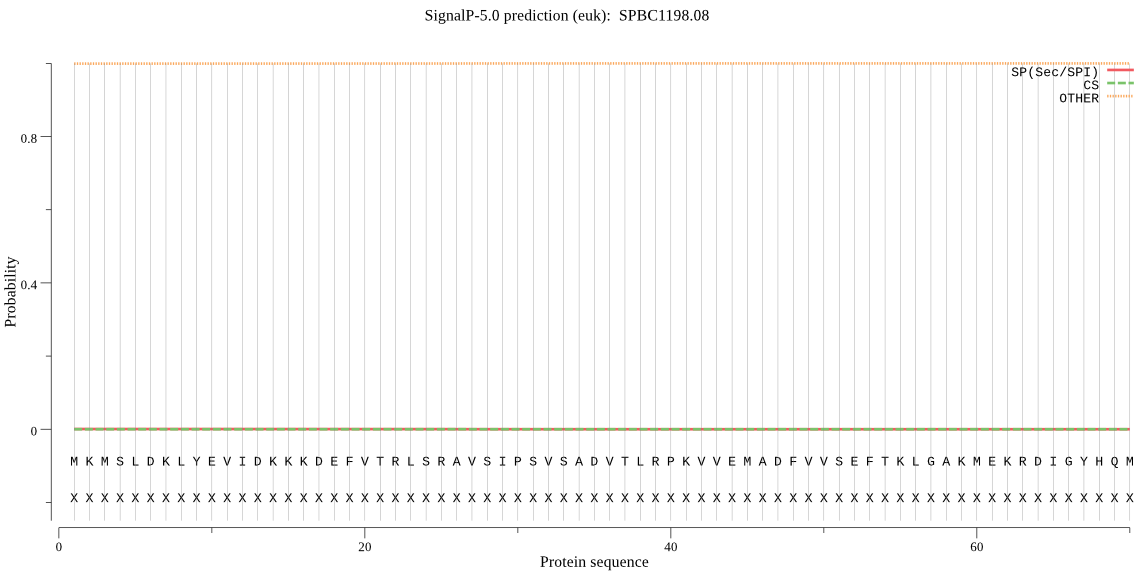
| SPBC1198.08 | OTHER | 0.998708 | 0.000467 | 0.000825 |
MKMSLDKLYEVIDKKKDEFVTRLSRAVSIPSVSADVTLRPKVVEMADFVVSEFTKLGAKM EKRDIGYHQMDGQDVPLPPIVLGQYGNDPSKKTVLIYNHFDVQPASLEDGWSTDPFTLTV DNKGRMFGRGATDDKGPLIGWISAIEAHKELGIDFPVNLLMCFEGMEEYGSEGLEDLIRA EAEKYFAKADCVCISDTYWLGTKKPVLTYGLRGVCYFNITVEGPSADLHSGVFGGTVHEP MTDLVAIMSTLVKPNGEILIPGIMDQVAELTPTEDSIYDGIDYTMEDLKEAVGADISIYP DPKRTLQHRWRYPTLSLHGIEGAFSGSGAKTVIPAKVIGKFSIRTVPNMESETVERLVKE HVTKVFNSLNSKNKLAFNNMHSGSWWISSPDHWHYDVGKKATERVYGITPDFVREGGSIP VTVTFEQSLKKNVLLLPMGRGDDGAHSINEKLDLDNFLKGIKLFCTYVHELASVSP
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| SPBC1198.08 | 106 S | VQPASLEDG | 0.995 | unsp | SPBC1198.08 | 276 S | PTEDSIYDG | 0.996 | unsp |