

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
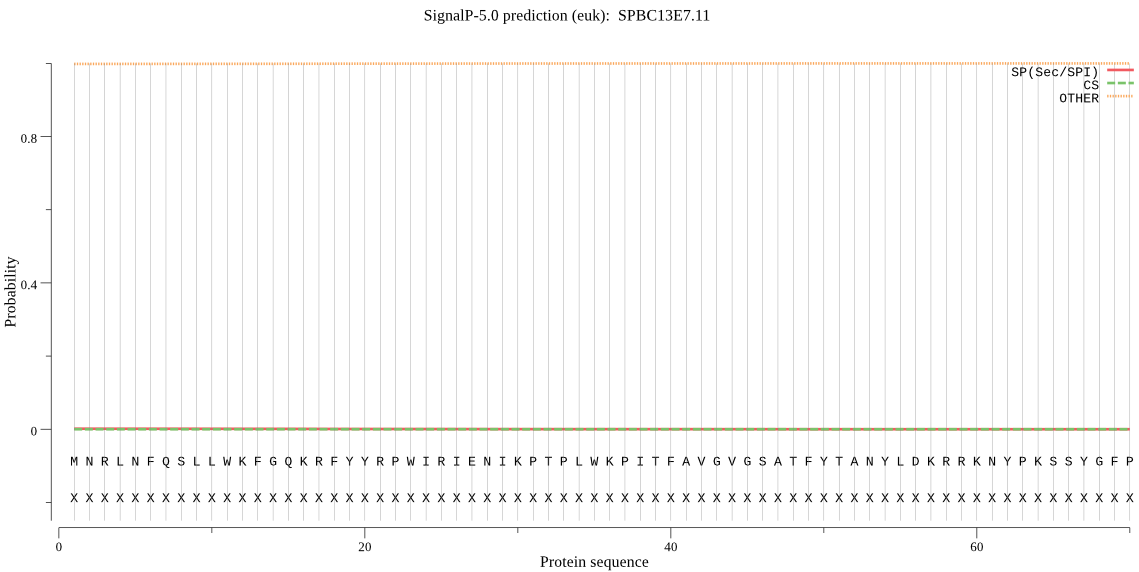
| SPBC13E7.11 | OTHER | 0.902518 | 0.000111 | 0.097371 |
MNRLNFQSLLWKFGQKRFYYRPWIRIENIKPTPLWKPITFAVGVGSATFYTANYLDKRRK NYPKSSYGFPIPQTSSRSLVLSIIGINVGVFALWRAPRFSHLNRFLQKYAVMNPIFINMP SMIVSAFSHQSGWHLLFNMVAFYSFAPAIVDVFGNNQFVAFYISSILFSNVASLLHHRLR FGTKVTPGSLGASGAIYAIAAATSYFFPNASVSIIFLPFIPIKIGVALLGLMAFDAWGLI SRGFSSFANFTLIDHAAHLGGGIFGWLYAKYGYSTYNRSTRPRPPSLSKPFFSRSVSF
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SPBC13E7.11 | S | 297 | 0.574 | 0.033 | SPBC13E7.11 | S | 295 | 0.560 | 0.025 | SPBC13E7.11 | S | 293 | 0.552 | 0.079 |
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SPBC13E7.11 | S | 297 | 0.574 | 0.033 | SPBC13E7.11 | S | 295 | 0.560 | 0.025 | SPBC13E7.11 | S | 293 | 0.552 | 0.079 |
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| SPBC13E7.11 | 286 S | PRPPSLSKP | 0.993 | unsp |