

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
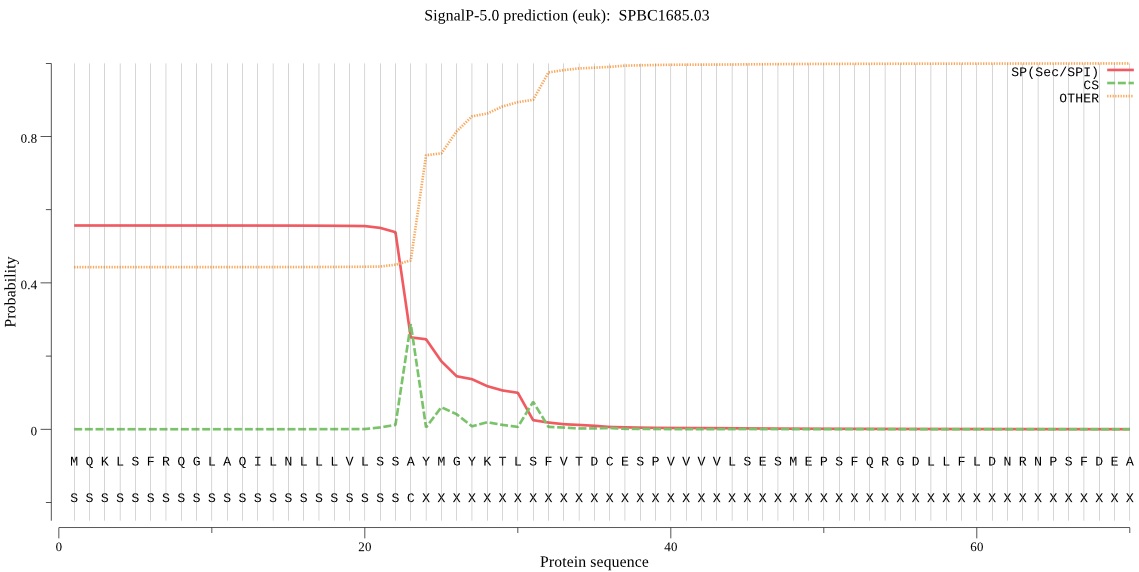
| SPBC1685.03 | SP | 0.084761 | 0.912936 | 0.002303 | CS pos: 31-32. TLS-FV. Pr: 0.3067 |
MQKLSFRQGLAQILNLLLVLSSAYMGYKTLSFVTDCESPVVVVLSESMEPSFQRGDLLFL DNRNPSFDEAKVPSVFEKIIYGSPVGIGDIVVYSLPDRPIPIVHRVVKLYESENQTHLIT KGDNNKIDDVAMFPKSINYLDRENHILGVVRGYFPYLGMITIWLTDYPILKYIMLGGLGL LTLIQKEEQ
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| SPBC1685.03 | 66 S | NRNPSFDEA | 0.996 | unsp |