

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
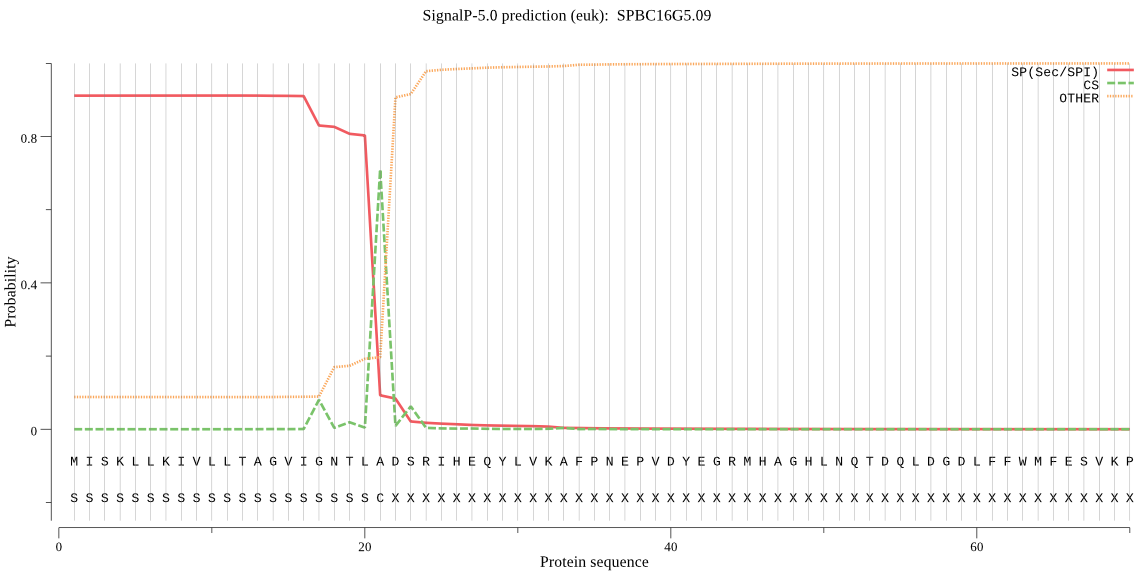
| SPBC16G5.09 | SP | 0.014567 | 0.984756 | 0.000678 | CS pos: 21-22. TLA-DS. Pr: 0.7744 |
MISKLLKIVLLTAGVIGNTLADSRIHEQYLVKAFPNEPVDYEGRMHAGHLNQTDQLDGDL FFWMFESVKPEYEHRSILWLNGGPGCSSEDGSLMEVGPFRLDDNNTFQLNPGRWDELGNL LFVDQPLGTGYSYSLAKDFQSNNEKMANDFSIFFEKFLEEFPERANDEWFIAGESFAGQY IPHIAAKLKEKNLVNLGGLAIGNGWINPLSHYETYLNYLVEKGMVDFESELGQYLHHSWA ECLLAFDKIGSGSGDLSKCESFLGDILYMVSKEPGKACMNMYDISLESTYPTCGMDWPYD LSYLTEFLSTREAMTSLNVNLEKVHDWEECNDDVALQYAREGIESSSKLIQDLVSTVPIL LFYGENDFLCNYLSGEKLTRSLEWNGAVGFQNQSAQPFYLPGYSDQPSGSYVSSRNLTFA RIVEASHMVPYDHPNEMKTLITAFFNNDFASLPSVPKPSPDLGNGNYKWLYLGLIPVALT IIILFSIYLCRRFGLFGLSKQRYQPISPTP
| ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|
| SPBC16G5.09 | T | 509 | 0.556 | 0.526 |
| ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|
| SPBC16G5.09 | T | 509 | 0.556 | 0.526 |
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| SPBC16G5.09 | 257 S | SGDLSKCES | 0.994 | unsp |