

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
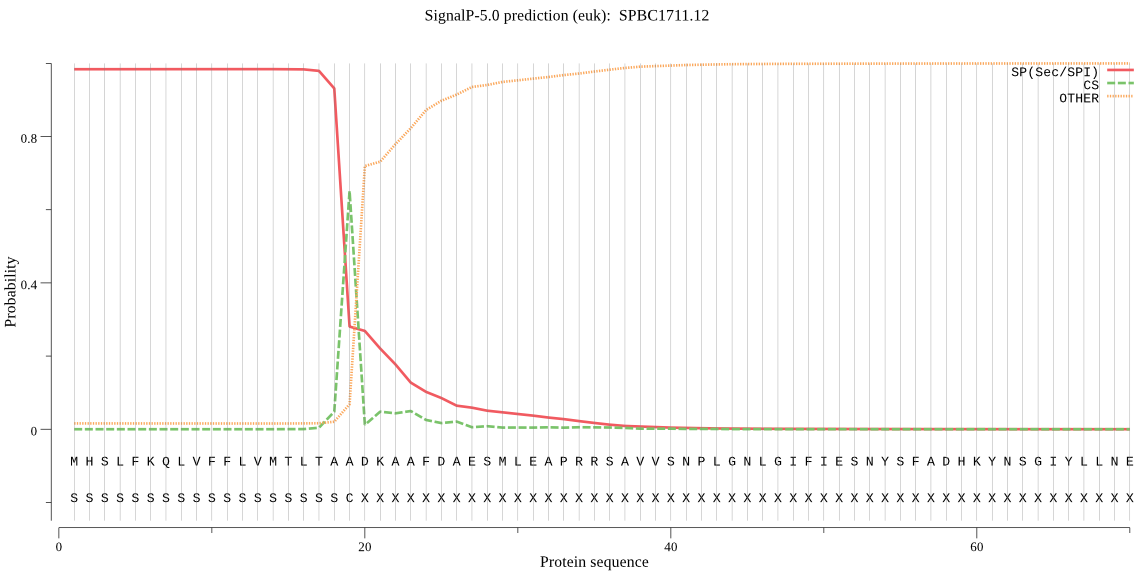
| SPBC1711.12 | SP | 0.001902 | 0.998062 | 0.000036 | CS pos: 19-20. TAA-DK. Pr: 0.7036 |
MHSLFKQLVFFLVMTLTAADKAAFDAESMLEAPRRSAVVSNPLGNLGIFIESNYSFADHK YNSGIYLLNESTRNHQELLVHGKSNKALTWITDSAFLYAREDNSSSSSILLFDVNNRSER IIYNHNSSISDIRIGEKNNHYRIVFSSVDNSLVKGPSNVHVYDHLFVRHWDRWNTGSRNT LYFIELDKKTENSNYFEISSEKAIDLLKETGLESPVEPFGGLSDFDSNYDKLVFVAKDPK LNPATQTKTVVYEINLNTRNLKSLSTAKGACSSPRLAKDGNHIAWLEMQTPQYESDQNQI MVYESESGAKKHIARHWDRSPSSIEWGVFKGGEPGLFAIAENYGKQILFFVSIFHHQVIP MTEEHSVSSISVPKSSSLWLTKSSLINPPYYAKINVETLNESVLLENNVGLSPTSYEEIW FPGTHGHRIHAWIVKPESFDKSKKYPVAVLIHGGPQGSWTDSWSTRWNPAVFANAGFIVF ALDPTGSTGYGQRFTDSIALDWGGKPYKDIELGVEYIKNHLSYADSEKMVALGASYGGYM INWIQGHPLGRQFRALVCHDGVFNTLNTFYNTEELYFSIHDFGGTPWENRVIYERWNPSN FVNYWATPELVIHSSKDYRLTESEGIAAFNVLQYKGIPSRLLVFEDENHWVIKPDNSLRW HKEVLSWILHYTKDCNANEDETF
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SPBC1711.12 | 214 S | TGLESPVEP | 0.992 | unsp | SPBC1711.12 | 214 S | TGLESPVEP | 0.992 | unsp | SPBC1711.12 | 214 S | TGLESPVEP | 0.992 | unsp | SPBC1711.12 | 323 S | RSPSSIEWG | 0.996 | unsp | SPBC1711.12 | 415 S | LSPTSYEEI | 0.996 | unsp | SPBC1711.12 | 614 S | LVIHSSKDY | 0.996 | unsp | SPBC1711.12 | 657 S | KPDNSLRWH | 0.991 | unsp | SPBC1711.12 | 55 S | ESNYSFADH | 0.99 | unsp | SPBC1711.12 | 128 S | NHNSSISDI | 0.994 | unsp |