

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
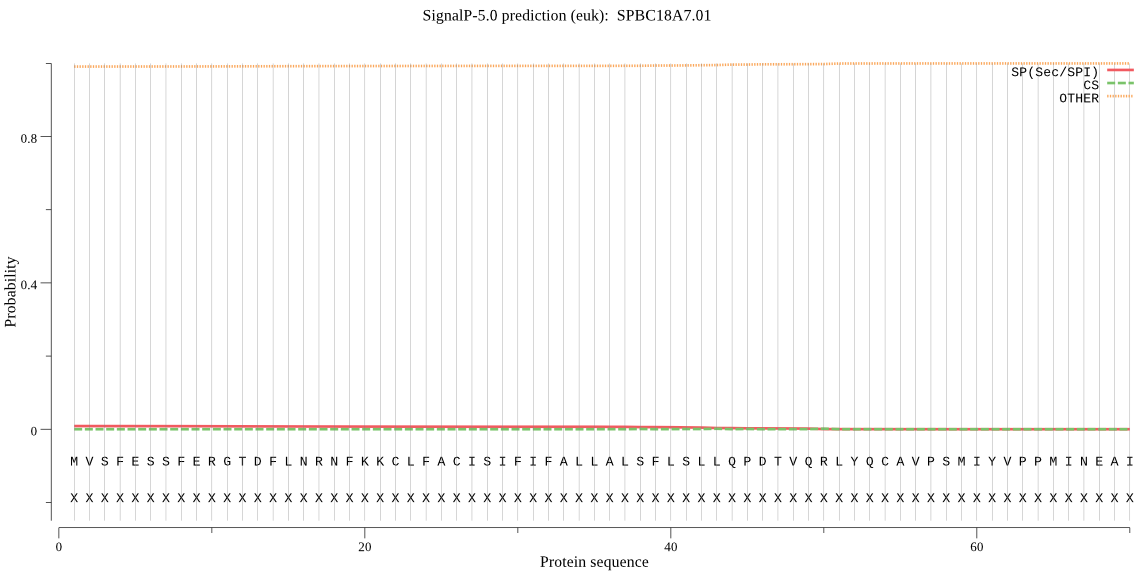
| SPBC18A7.01 | OTHER | 0.982866 | 0.012862 | 0.004272 |
MVSFESSFERGTDFLNRNFKKCLFACISIFIFALLALSFLSLLQPDTVQRLYQCAVPSMI YVPPMINEAISIQHEEFNNRRRRLSAALREDKLDALIMEPTVSMDYFANITTGSWGLSER PFLGIIFSDDEPYPGDVASRIYFLVPKFELPRAKELVGKNIDAKYITWDEDENPYQVLYD RLGPLKLMIDGTVRNFIAQGLQYAGFTTFGVSPRVASLREIKSPAEVDIMSRVNIATVAA IRSVQPCIKPGITEKELAEVINMLFVYGGLPVQESPIVLFGERAAMPHGGPSNRRLKKSE FVLMDVGTTLFGYHSDCTRTVLPHGQKMTERMEKLWNLVYDAQTAGIQMLSHLSNTSCAE VDLAARKVIKDAGYGEYFIHRLGHGLGLEEHEQTYLNPANKGTPVQKGNVFTVEPGIYIP DEIGIRIEDAVLASDVPILLTNFRAKSPYEP
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SPBC18A7.01 | 447 S | FRAKSPYEP | 0.997 | unsp | SPBC18A7.01 | 447 S | FRAKSPYEP | 0.997 | unsp | SPBC18A7.01 | 447 S | FRAKSPYEP | 0.997 | unsp | SPBC18A7.01 | 212 S | TFGVSPRVA | 0.991 | unsp | SPBC18A7.01 | 217 S | PRVASLREI | 0.996 | unsp |