

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
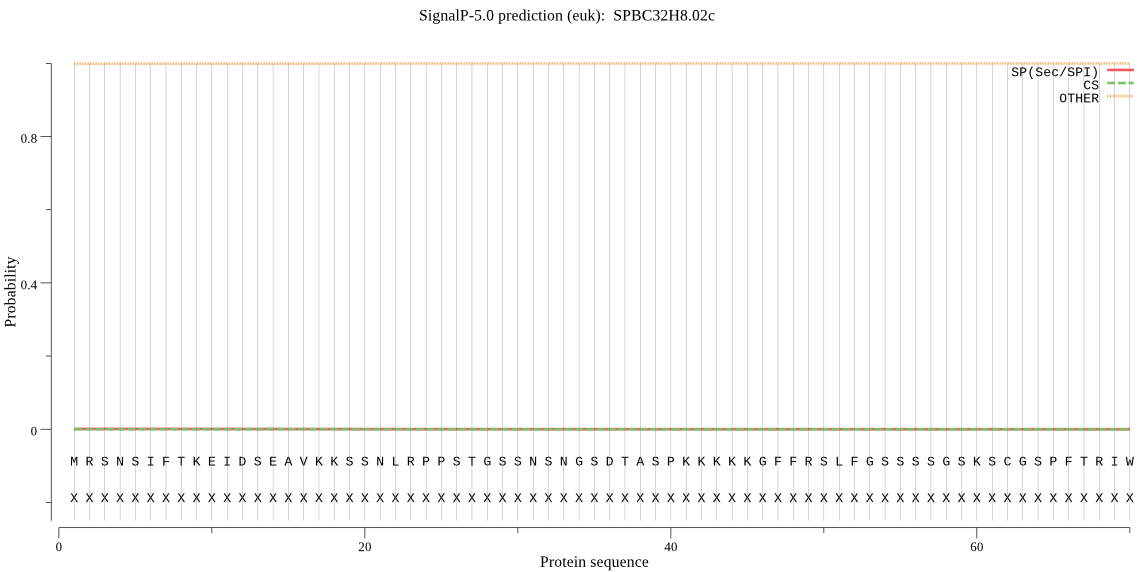
| SPBC32H8.02c | OTHER | 0.999938 | 0.000009 | 0.000053 |
MRSNSIFTKEIDSEAVKKSSNLRPPSTGSSNSNGSDTASPKKKKKGFFRSLFGSSSSGSK SCGSPFTRIWLEYFEVSLRKNDVDHFRPGYWILDTNIDFFYEIMLRQVLLKRPKEESQQI YLLRPAMVFFLAQAPNPLEIESALPPAMFDASFIFLPINDTNECGIESGSHWSLLVVSVE KGLGWYYDSMSNGNTNDCNLAIKNLGILLKKEFRVRHMKTPQQINDCDCGLHVCENTRIL MYRLLQKPYVPKVDMNLDHSVVDSVRLRKALMEVITSLLAAYGSKVPKPSETHTDPEKDK KIFSKICKTEELLELPTLSAVTSDSAQPHSLPASMPSSQPQSRSESLPLTHPNSEPNPKL DSQPNSSPVRRPSLIKVKTASTSVLPTSILQRPPSIVPRPETAAIQHTQQSIEIH
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SPBC32H8.02c | 342 S | SQPQSRSES | 0.993 | unsp | SPBC32H8.02c | 342 S | SQPQSRSES | 0.993 | unsp | SPBC32H8.02c | 342 S | SQPQSRSES | 0.993 | unsp | SPBC32H8.02c | 373 S | VRRPSLIKV | 0.993 | unsp | SPBC32H8.02c | 39 S | SDTASPKKK | 0.995 | unsp | SPBC32H8.02c | 260 S | NLDHSVVDS | 0.997 | unsp |