

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
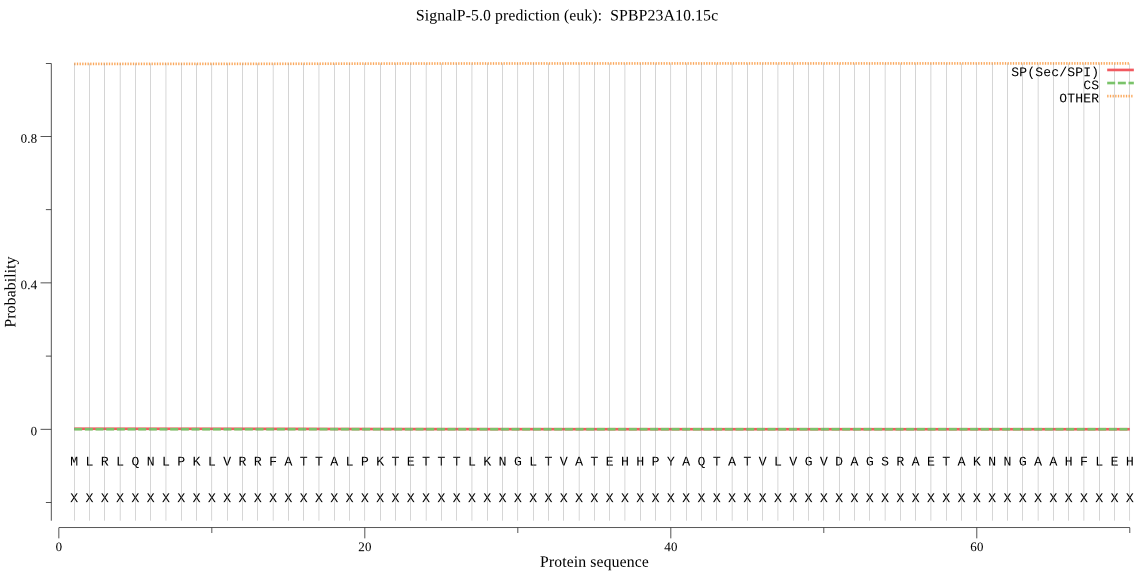
| SPBP23A10.15c | mTP | 0.012326 | 0.000044 | 0.987630 | CS pos: 14-15. RRF-AT. Pr: 0.8188 |
MLRLQNLPKLVRRFATTALPKTETTTLKNGLTVATEHHPYAQTATVLVGVDAGSRAETAK NNGAAHFLEHLAFKGTKNRSQKALELEFENTGAHLNAYTSREQTVYYAHAFKNAVPNAVA VLADILTNSSISASAVERERQVILREQEEVDKMADEVVFDHLHATAYQGHPLGRTILGPK ENIESLTREDLLQYIKDNYRSDRMIISSAGSISHEELVKLAEKYFGHLEPSAEQLSLGAP RGLKPRFVGSEIRARDDDSPTANIAIAVEGMSWKHPDYFTALVMQAIIGNWDRAMGASPH LSSRLSTIVQQHQLANSFMSFSTSYSDTGLWGIYLVTENLGRIDDLVHFTLQNWARLTVA TRAEVERAKAQLRASLLLSLDSTTAIAEDIGRQLLTTGRRMSPQEVDLRIGQITEKDVAR VASEMIWDKDIAVSAVGSIEGLLDYNRIRSSISMNRW
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SPBP23A10.15c | 272 S | VEGMSWKHP | 0.995 | unsp | SPBP23A10.15c | 272 S | VEGMSWKHP | 0.995 | unsp | SPBP23A10.15c | 272 S | VEGMSWKHP | 0.995 | unsp | SPBP23A10.15c | 324 S | SFSTSYSDT | 0.997 | unsp | SPBP23A10.15c | 402 S | GRRMSPQEV | 0.998 | unsp | SPBP23A10.15c | 80 S | TKNRSQKAL | 0.995 | unsp | SPBP23A10.15c | 213 S | AGSISHEEL | 0.995 | unsp |