

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
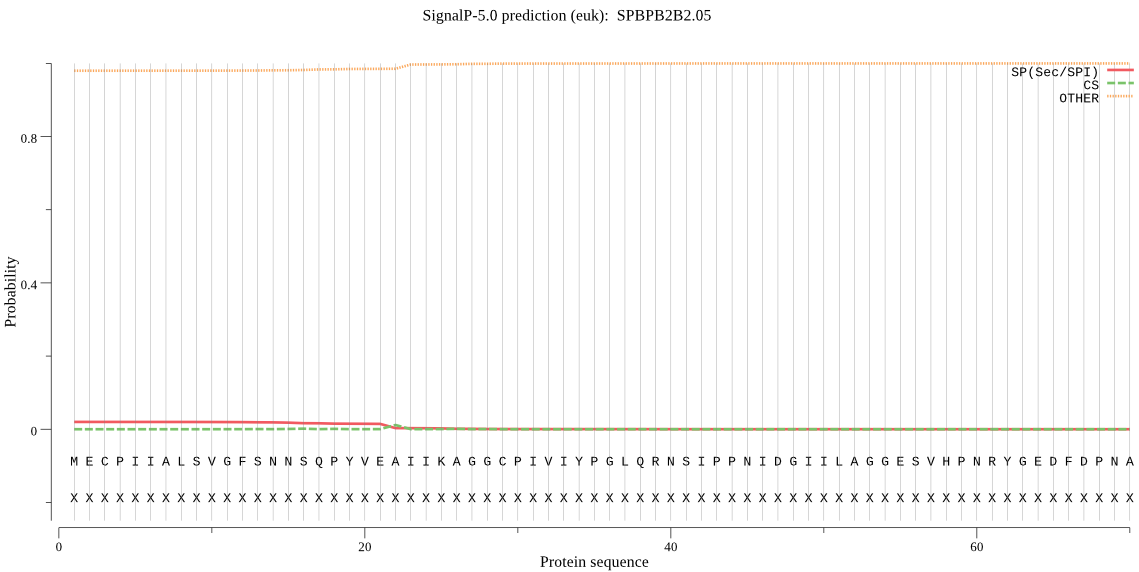
| SPBPB2B2.05 | OTHER | 0.922950 | 0.075075 | 0.001976 |
MECPIIALSVGFSNNSQPYVEAIIKAGGCPIVIYPGLQRNSIPPNIDGIILAGGESVHPN RYGEDFDPNAPKSVDVIRDSTEWGMIDFALKKKIPILGICRGCQVLNVYFGGSLYQNVSS CGFRDIHRPSKPRHYLAHKVMAKPGKLKNILGSNVIDVNSIHDQGIKTLGMGLQSTVISD DGLCEGIESKDGLIIGVQWHPEAIIDKQPHSLKLFQYFINRSKWHMKQSNIFSNVPHESS FYRNSIISIPIAP
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| SPBPB2B2.05 | 80 S | VIRDSTEWG | 0.997 | unsp | SPBPB2B2.05 | 130 S | IHRPSKPRH | 0.991 | unsp |