

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
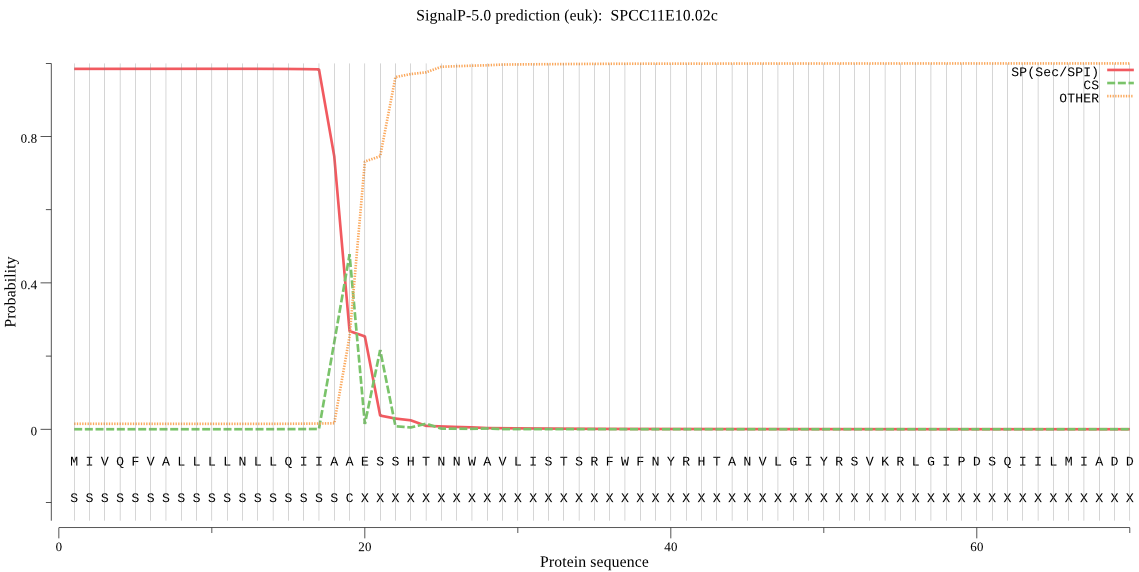
| SPCC11E10.02c | SP | 0.006900 | 0.992989 | 0.000111 | CS pos: 19-20. IAA-ES. Pr: 0.7231 |
MIVQFVALLLLNLLQIIAAESSHTNNWAVLISTSRFWFNYRHTANVLGIYRSVKRLGIPD SQIILMIADDYACNSRNLFPGTVFDNADRALDLYGEEIEIDYKGYEVTVEAFIRLLTERV PENTPASKRLLTNERSNILIYMTGHGGDGFIKFQDAEELSSEDLADAIEQIHQHKRYNEI LFMVDTCQANSLYTKIYSPNVLAIGSSEVGTSSYSHHADIDIGVAVIDRFTFSNLEFLEN RVDSKSKLTMQDLINSYNPYEIHSTPGVQPINLRRSPDDILITDFFGNVRDIELHSEKIN WMLPGENTTKPSIKRNSFVFQAQNDMQDDGKGFGISNLKSFLPPTRELKYKKHPISRIIS AVVCISFSIGFPYYASKYLK
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SPCC11E10.02c | 276 S | NLRRSPDDI | 0.998 | unsp | SPCC11E10.02c | 276 S | NLRRSPDDI | 0.998 | unsp | SPCC11E10.02c | 276 S | NLRRSPDDI | 0.998 | unsp | SPCC11E10.02c | 160 S | AEELSSEDL | 0.995 | unsp | SPCC11E10.02c | 244 S | NRVDSKSKL | 0.99 | unsp |