

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
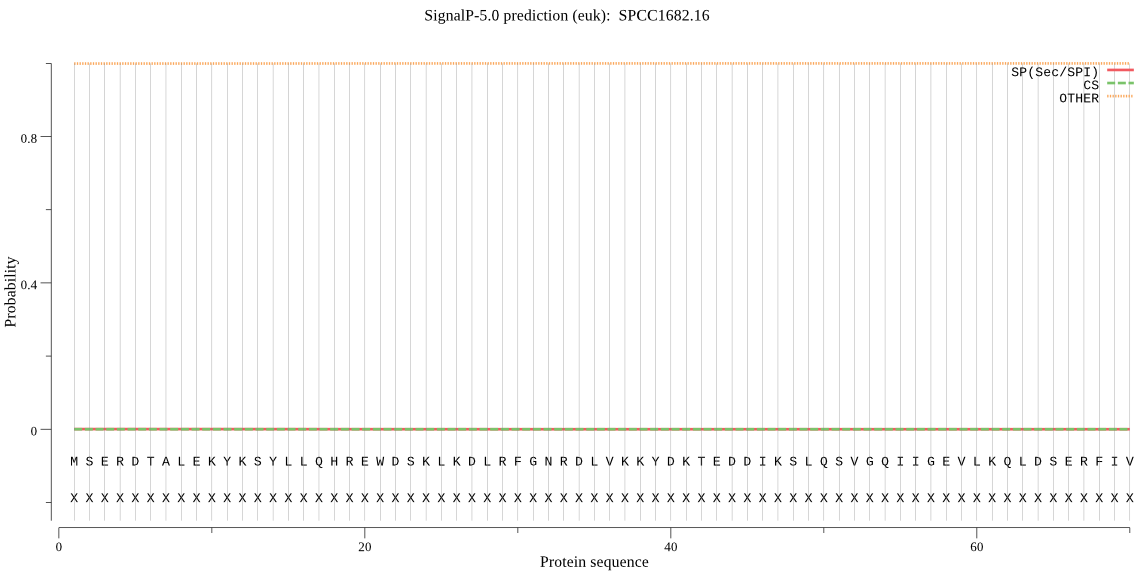
| SPCC1682.16 | OTHER | 0.999953 | 0.000024 | 0.000023 |
MSERDTALEKYKSYLLQHREWDSKLKDLRFGNRDLVKKYDKTEDDIKSLQSVGQIIGEVL KQLDSERFIVKASSGPRYVVGCRNNVDQSHLVQGVRVSLDMTTLTIMRILPREVDPLVYN MSIEDPGDISFAGVGGLNEQIRELREVIELPLKNPELFLRVGIKPPKGVLLYGPPGTGKT LLARAVAASLGVNFLKVVSSAIVDKYIGESARIIREMFGYAKEHEPCVIFMDEIDAIGGR RFSEGTSADREIQRTLMELLNQMDGFDYLGQTKIIMATNRPDTLDPALLRPGRLDRKIEI PLPNEVGRMEILKIHLEKVSKQGEIDYEALVKLTDGTNGADLRNVVTEAGFIAIKEDRDY VIQSDLMSAARKVADLKKLEGTIDYQKL
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| SPCC1682.16 | 243 S | GRRFSEGTS | 0.994 | unsp |