

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| SPCC757.05c | SP | 0.013303 | 0.986340 | 0.000357 | CS pos: 25-26. VLA-GP. Pr: 0.9788 |
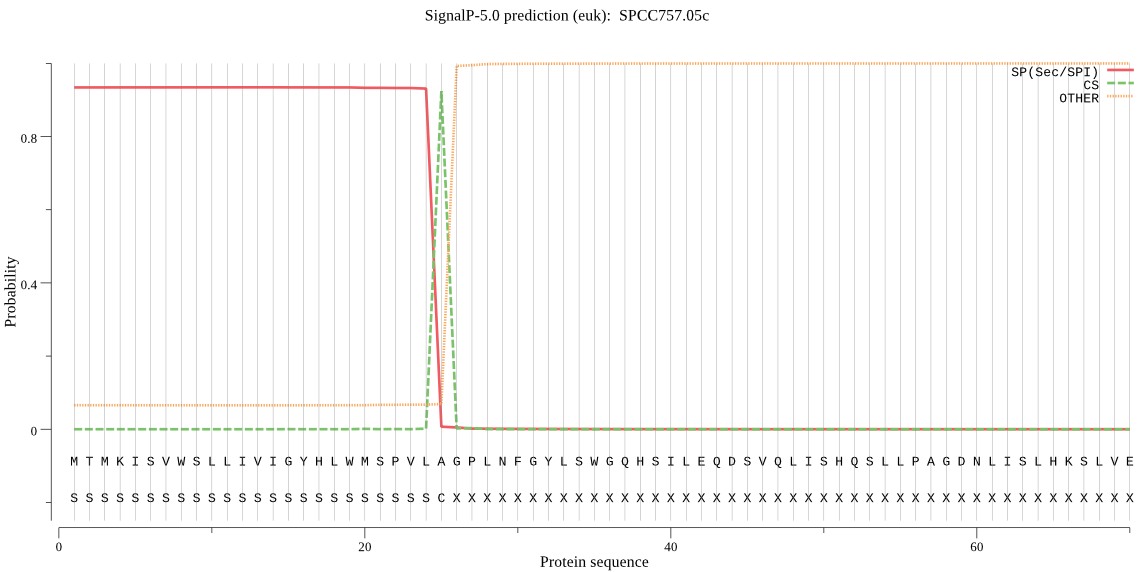
MTMKISVWSLLIVIGYHLWMSPVLAGPLNFGYLSWGQHSILEQDSVQLISHQSLLPAGDN LISLHKSLVEIESLGGNEVNVSDFLKSYLESKGLTVELQRVSSNPTVRDNVYAYLGSQRN TKVVLTSHIDTVNPFLPYYIEGDKIHGRGSCDAKSSVAAQIFAMLELMSEGKVQEGDLSL LFVVGEEIDGIGMKTVANKLNADWEVAIFGEPTENKLGVGHKGNFRFDVYAHGKACHSGY PQEGFSAIEFLLRQCVKLMDTDLPKSKLLGPSTINIGTIEGGAAANILAAEAKAEVFIRV AEDIETIRDIAEDLFDTEHSEIKVIQYSPPQYLDYDIPGMDTVVLAYATDIPYLSDRKMK IYLFGPGSIREAHGPNEYVTFSQLFEGLNGYKRMVMYNLR
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| SPCC757.05c | 127 S | VVLTSHIDT | 0.996 | unsp |