

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
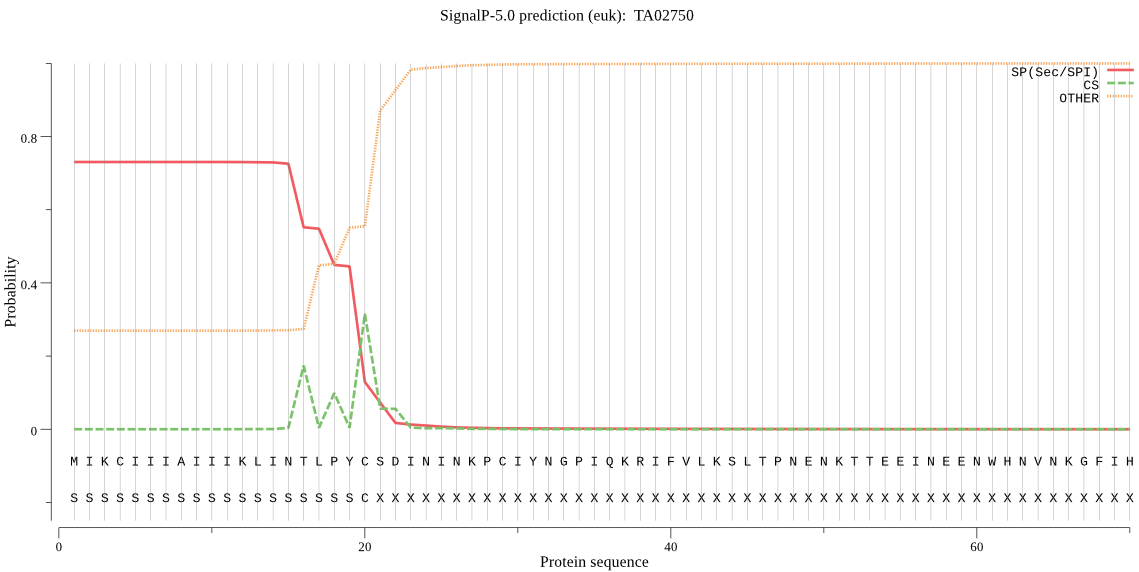
| TA02750 | SP | 0.062423 | 0.937189 | 0.000388 | CS pos: 16-17. INT-LP. Pr: 0.7166 |
MIKCIIIAIIIKLINTLPYCSDININKPCIYNGPIQKRIFVLKSLTPNENKTTEEINEEN WHNVNKGFIHYFYRSSVTGSGPTASNGPKEAVTEDSSTVGPNTVTEENTSTPGKGANSMP MECNSSNIEEYPSGVGRGPHSVTGKTKLINIKLHNGILIPHLRHVQYALNMGVGTPKQEI NPIIDTGSTNTWVISQNCISNTCEGVASFNSKKSETFNPINEGLKIKFGTGIIKGVLGID NITIGNDIIKQQIFGLVNEENSNIFNVIKFQGIIGLGFPKLAFDHHTSLYDNYSKQINAD LIFSLYFSNQYSYLMMGGIDKRFYKGDLYMLPVIRELYWEIKLYELWIGNIKLCCNNESY IIFDSGTSFNTMPHTEFLLFKQYIKPKYCNGLENIYEEYPIIKYKLEGGIEINIEPQEYI FLHKDKCRIAYMQIDVPSSYGRAFILGTQAFMTHYYTVYQRQPPMVRYGACSNSNSTKTG SLTPQRGFLTSERGLFSYTSYLPLVVIYLQYLVLFPGSI
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TA02750 | T | 98 | 0.632 | 0.660 | TA02750 | T | 105 | 0.618 | 0.070 | TA02750 | T | 93 | 0.607 | 0.116 | TA02750 | T | 103 | 0.591 | 0.021 | TA02750 | T | 111 | 0.553 | 0.069 | TA02750 | S | 97 | 0.550 | 0.021 | TA02750 | T | 109 | 0.519 | 0.039 | TA02750 | S | 96 | 0.517 | 0.031 |
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TA02750 | T | 98 | 0.632 | 0.660 | TA02750 | T | 105 | 0.618 | 0.070 | TA02750 | T | 93 | 0.607 | 0.116 | TA02750 | T | 103 | 0.591 | 0.021 | TA02750 | T | 111 | 0.553 | 0.069 | TA02750 | S | 97 | 0.550 | 0.021 | TA02750 | T | 109 | 0.519 | 0.039 | TA02750 | S | 96 | 0.517 | 0.031 |
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| TA02750 | 439 S | DVPSSYGRA | 0.994 | unsp |