

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
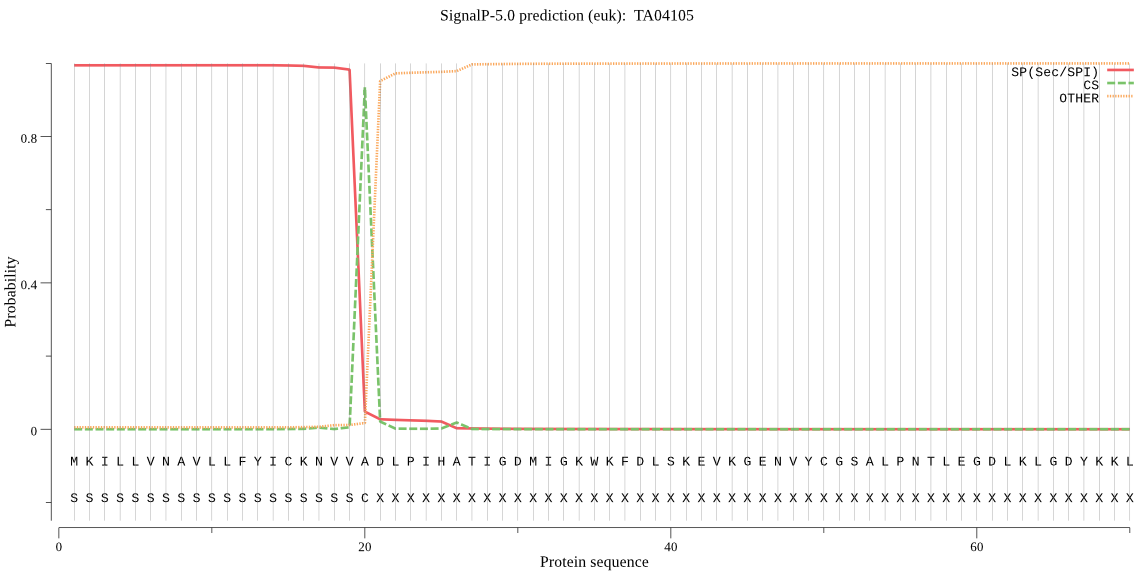
| TA04105 | SP | 0.001943 | 0.998024 | 0.000034 | CS pos: 20-21. VVA-DL. Pr: 0.9684 |
MKILLVNAVLLFYICKNVVADLPIHATIGDMIGKWKFDLSKEVKGENVYCGSALPNTLEG DLKLGDYKKLMTNNYELGNTLELELTLDPIQYINVSKAKNREKWISLAVKSYGNTVGHWT AIYDQGFRITLNNGYDLLLYFYFEKIEENKYATDPTRTLIGWASKRDGNNFTHYCCYGEK KNSGENNVYNTVDTTETGYERFARDLIRVVNGKKNFHPVTNKGWSEYNVSYFCKCSKESR DNGNSNLPKNFSWDRFDNIQIVDQKGCGGCYTIASLFVLHSRFLIRIREMLKTPEYKSDA RLLHLEKVLTEKNFNINMSLSCIPYNQGCKGGFPVNVGKFAQEFGLVLDDEKPEEIVDNL SCAPKGDNLRLYASNVEYIGGCYECTRCSGETLIMSEIMENGPVVAGIDGEHIRKYKDSV INPSKEDLRKHRGLCEFNEKFLSGLEFTTHAVVLVGWGETDEGFKFWVARNSWGKNWGDG GFFKIVRGINAFGIESEAVVLDPDVEKVLKNTTEQPINSHELTQTLL
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| TA04105 | 183 S | EKKNSGENN | 0.995 | unsp | TA04105 | 424 S | VINPSKEDL | 0.997 | unsp |