

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| TA04310 | OTHER | 0.925746 | 0.029573 | 0.044682 |
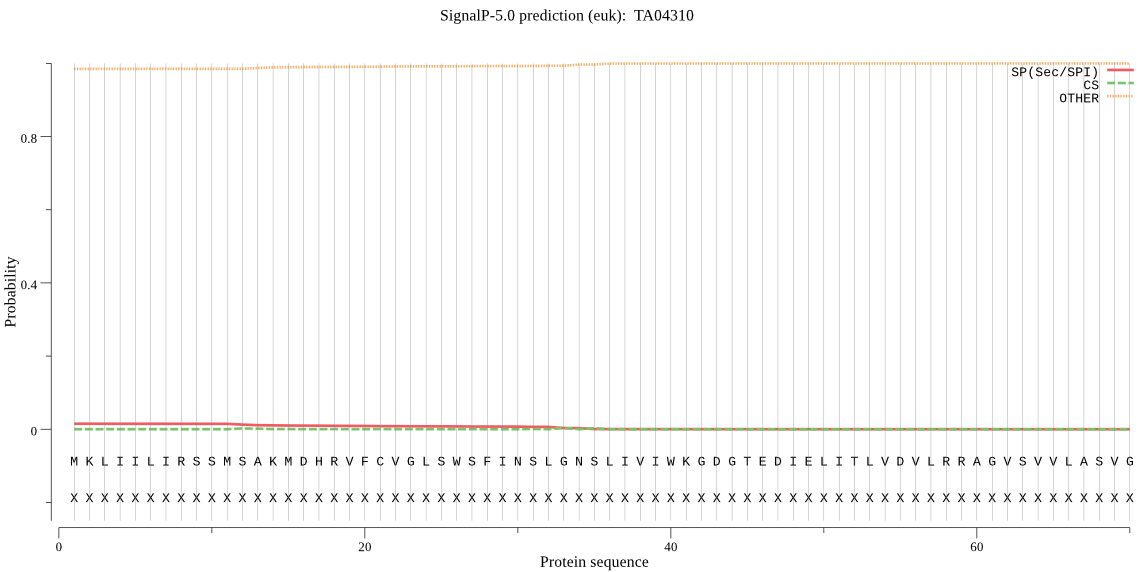
MKLIILIRSSMSAKMDHRVFCVGLSWSFINSLGNSLIVIWKGDGTEDIELITLVDVLRRA GVSVVLASVGDSVNLVLAHGTKLTADDKVVNLTQKTFDLIAVPGGLVGATNCANNVTLIR MLKEQKSNGRLYAAICASPALVFGDCGLLDDKTSAVAFPGFENKLPLVGTGRVHVSNNCG IFRYVTSQGPGTALEFALKLVELLCGVEAKNKLTKSMLLHPSININSLYDLLICMSSNNN RLKMAIKVTEKGTK
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| TA04310 | 12 S | RSSMSAKMD | 0.995 | unsp | TA04310 | 68 S | VVLASVGDS | 0.994 | unsp |