-
Computed_GO_Component_IDs:
-
Computed_GO_Components:
-
Computed_GO_Function_IDs:
-
Computed_GO_Functions:
-
Computed_GO_Process_IDs:
-
Computed_GO_Processes:
-
Curated_GO_Component_IDs:
-
Curated_GO_Components:
-
Curated_GO_Function_IDs:
-
Curated_GO_Functions:
-
Curated_GO_Processes:
No Results
-
Fasta :-
>TA05610
MVISGVGSVKVKVNTNTTNSTKDSTNSTKDISSTAGTIGASTVTEGKGANSMGMECTTNN
TKEAPFGAGTKDITGREPGTVTEDTVMELNEILKNDTSAASIAATSLLISNTYLNSKDIF
DNGKCFIGGLKPSEMYLFNNKSQTELLYNSSVTVLGQTDTIPPNSTTNPLTNSSSTDPST
VGASTVTEEKNSNEIAVVTKSGESGTFLDTVGIEGAPIRAVGEVVNTKDSENICTVGAST
VTDTVTEEKIINSYNLWSHENIEDKLTNNKGYHDLGYKYNKTMESKDIESELLNITNVLP
YDGNPKIRSRYYVTSASHLFSVFNFFKYAHLLDDSFDTNSKQIENINDLHYLSHIVLRVW
RSKSDKGYFNRLEILVSSGAKDGFGQNYQLLEKNAKNQKNNYKKYIQKLQLHKFSKFCNQ
CYIPSNLNNTNGTTSTGTVTDTTGTMGTTGVNVMNINVLDKYNLMYENIKEKKFKCYNCF
LKEYKTLYNLQNFTTDNFTTADLGFGSPNIRPNSVDIGLNTTDIGLNTPDIGLNTPNNNL
DKFNTDNNLDKFNTDNFSTLDIGFKPDIGLSTQDIGLNRGMNNGVKMVPPYCELNTLVTM
NRNFGLDRLKDYIQKAYHNILLKN
- Download Fasta
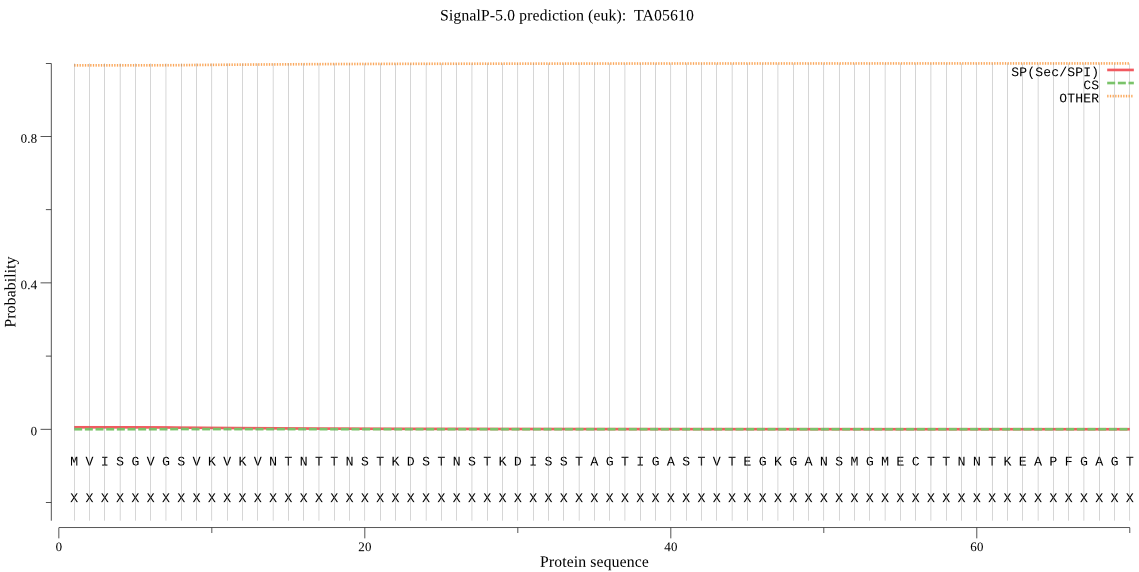
-
MitoProt II - v1.101
File : /home/rajan/sadaf/4480_mitoprot/test/TA05610.fa
Sequence name : TA05610
Sequence length : 624
VALUES OF COMPUTED PARAMETERS
Coef20 : 4.099
CoefTot : -0.905
ChDiff : -8
ZoneTo : 22
KR : 3
DE : 0
CleavSite : 0
HYDROPHOBIC SCALE USED
GES KD GVH1 ECS
H17 : 1.194 1.071 0.110 0.427
MesoH : -0.493 0.010 -0.408 0.135
MuHd_075 : 11.340 10.000 3.712 2.585
MuHd_095 : 12.419 14.827 4.070 3.687
MuHd_100 : 12.022 15.000 4.054 3.572
MuHd_105 : 12.458 10.986 3.973 2.366
Hmax_075 : 11.900 9.900 2.009 3.442
Hmax_095 : 6.125 12.200 0.044 4.050
Hmax_100 : 5.800 16.700 0.133 4.250
Hmax_105 : 3.000 13.900 -1.475 3.520
CLASS NOT-MITO MITO(/CHLORO)
DFM : 0.9423 0.0577
DFMC : 0.9001 0.0999
-
Fasta :-
>TA05610
ATGGTAATAAGTGGGGTAGGCTCAGTTAAGGTTAAGGTTAATACTAATACTACTAACAGT
ACTAAAGATAGTACCAACAGTACTAAGGATATTAGTAGTACTGCTGGTACCATTGGAGCA
AGCACCGTTACTGAGGGAAAGGGAGCTAATTCTATGGGTATGGAGTGTACTACTAATAAT
ACTAAGGAAGCCCCTTTCGGGGCTGGTACTAAGGATATTACTGGCCGTGAACCTGGCACC
GTAACGGAAGACACCGTAATGGAATTAAATGAGATATTAAAAAATGATACAAGTGCAGCA
TCAATAGCGGCAACAAGTTTATTAATATCAAATACATATTTAAATTCTAAAGATATATTT
GATAATGGTAAATGTTTTATTGGTGGTTTAAAACCTTCTGAAATGTATCTTTTTAATAAT
AAATCACAAACTGAATTATTATATAATTCCTCAGTAACGGTGCTTGGTCAAACAGATACT
ATTCCTCCTAACAGTACTACTAATCCCCTAACTAACAGTAGTAGTACTGATCCTAGTACC
GTAGGAGCAAGCACCGTTACGGAGGAGAAAAATTCTAATGAAATTGCTGTTGTCACTAAA
TCTGGGGAGTCAGGTACTTTTTTGGATACTGTAGGTATTGAGGGAGCCCCTATCAGGGCT
GTTGGTGAGGTTGTTAATACTAAGGACAGTGAGAATATTTGTACAGTTGGAGCAAGCACC
GTAACGGACACCGTAACGGAGGAAAAAATAATAAATTCATATAATTTATGGTCACATGAG
AATATAGAAGATAAATTAACAAATAATAAAGGATATCATGATTTAGGATATAAATATAAT
AAAACAATGGAAAGTAAAGATATAGAATCGGAATTATTAAATATAACAAATGTATTACCA
TATGATGGAAATCCAAAAATCAGATCACGTTATTATGTAACATCAGCATCACATTTATTT
AGTGTATTTAATTTCTTTAAATATGCACATTTACTTGATGATTCATTTGATACTAATTCC
AAACAAATTGAAAATATTAATGATTTACATTATTTATCACATATTGTATTACGAGTATGG
CGTAGTAAATCTGATAAAGGTTATTTTAATAGATTAGAAATACTTGTCAGTTCCGGTGCT
AAAGATGGTTTTGGACAAAATTATCAATTATTAGAAAAAAATGCTAAAAATCAAAAAAAC
AATTATAAAAAATATATACAAAAATTACAATTACACAAATTTTCCAAATTCTGTAATCAA
TGTTATATACCATCCAATCTTAATAATACCAACGGTACTACTAGTACTGGTACGGTGACT
GATACTACGGGTACTATGGGTACTACGGGTGTTAATGTTATGAATATAAATGTATTGGAT
AAATATAATTTAATGTATGAAAATATAAAAGAAAAGAAATTTAAATGTTATAATTGTTTT
TTAAAAGAATATAAAACTTTATATAATCTTCAAAATTTTACTACTGATAATTTTACTACT
GCAGATTTAGGATTTGGTAGTCCAAATATACGACCAAATAGTGTGGATATAGGATTAAAT
ACTACAGATATTGGATTAAATACACCGGATATAGGATTAAATACACCAAATAATAATTTG
GATAAATTTAATACAGATAATAATTTGGATAAATTTAATACAGATAATTTTAGTACACTA
GATATAGGATTTAAACCGGATATAGGATTAAGTACACAGGATATAGGATTGAATAGAGGA
ATGAATAATGGAGTGAAGATGGTACCACCATATTGTGAATTAAATACATTAGTAACAATG
AATAGAAATTTTGGATTAGATCGTTTAAAAGATTATATACAAAAAGCATATCATAATATA
TTACTCAAAAATTAA
- Download Fasta
|
ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore |
|
TA05610 | T | 171 | 0.659 | 0.056 | TA05610 | T | 166 | 0.636 | 0.511 | TA05610 | T | 167 | 0.628 | 0.069 | TA05610 | T | 176 | 0.616 | 0.072 | TA05610 | T | 180 | 0.580 | 0.076 | TA05610 | S | 173 | 0.572 | 0.039 | TA05610 | S | 174 | 0.552 | 0.025 | TA05610 | S | 175 | 0.543 | 0.027 | TA05610 | S | 165 | 0.527 | 0.022 | TA05610 | T | 28 | 0.513 | 0.078 | TA05610 | T | 185 | 0.505 | 0.024 | TA05610 | T | 187 | 0.501 | 0.076 |
|
ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore |
|
TA05610 | T | 171 | 0.659 | 0.056 | TA05610 | T | 166 | 0.636 | 0.511 | TA05610 | T | 167 | 0.628 | 0.069 | TA05610 | T | 176 | 0.616 | 0.072 | TA05610 | T | 180 | 0.580 | 0.076 | TA05610 | S | 173 | 0.572 | 0.039 | TA05610 | S | 174 | 0.552 | 0.025 | TA05610 | S | 175 | 0.543 | 0.027 | TA05610 | S | 165 | 0.527 | 0.022 | TA05610 | T | 28 | 0.513 | 0.078 | TA05610 | T | 185 | 0.505 | 0.024 | TA05610 | T | 187 | 0.501 | 0.076 |
|
ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method |
|
TA05610 | 204 S | KSGESGTFL | 0.991 | unsp | TA05610 | 204 S | KSGESGTFL | 0.991 | unsp | TA05610 | 204 S | KSGESGTFL | 0.991 | unsp | TA05610 | 364 S | WRSKSDKGY | 0.992 | unsp | TA05610 | 20 S | NTTNSTKDS | 0.998 | unsp | TA05610 | 27 S | DSTNSTKDI | 0.998 | unsp |


