

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
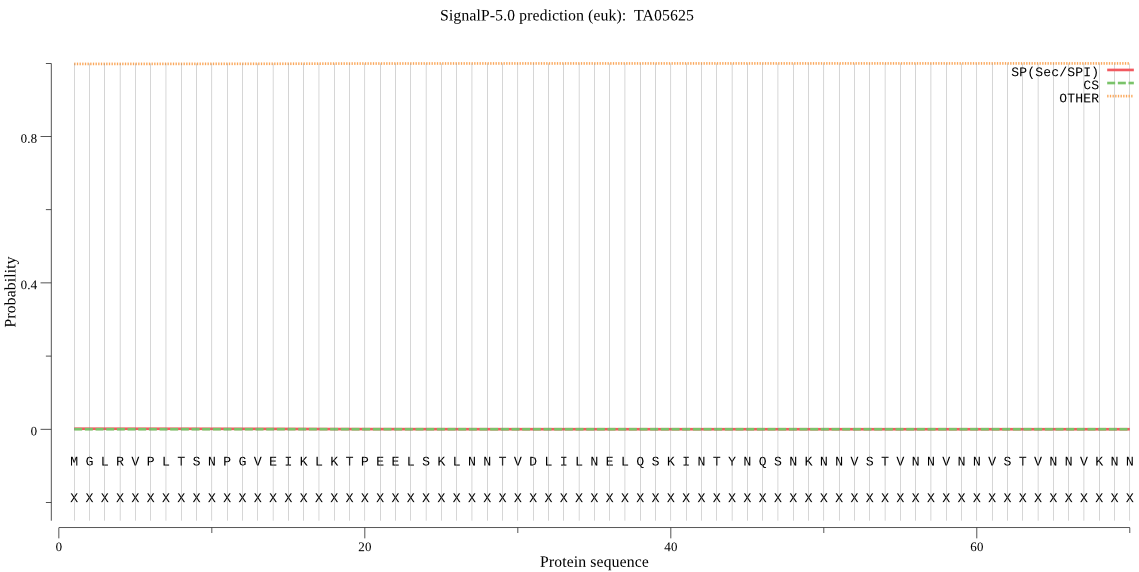
| TA05625 | OTHER | 0.999863 | 0.000029 | 0.000108 |
MGLRVPLTSNPGVEIKLKTPEELSKLNNTVDLILNELQSKINTYNQSNKNNVSTVNNVNN VSTVNNVKNNVSTVNNVESSVPITKVTTNSVTKDVNMKNLEEVDIKMLMNEFENFKELKK MLDKGYEFSGINRKVQLKPQFINIPMNSSVTVLGHTATNGTEVTSTNNSTNSTNSTNSTK DPTGASTVTEGKGANSMPMKCTTTNINEPSNINIESYSNTPTNTITTPTNTTHNHTTSNH ITSTDTPINTLTTPSNTITTPTTSPNPITTNITNNNSINRMNEIDKKLDSVLIVAKWGGD LTEVGKLQAEDLGRRFRETLYPADCSGLIRLHSTFRHDFKIYSSDEGRCQIXSASFTKGI LELEGDLTPILVTMTIRNKRAYELLNDTTISKQRTRCKMLLNQLISTFGTKLYTQILKQL TEYDRYYYHKAIQDSEYKLQDFPQLYNLIIKYIKNIKNELIWNSLYSYQYNFFINKLNNI INRWYNLLNKFASVTVLAHTDSNGPDINTTTNSTNYIGTIGPSTVTEENTSTPGKGANSM PMECNSSNIEEYPSGVGRGPHSVTGKTKLINIKLHNGILIPHLRHVQYALNMGVGTPKQE INPIIDTGSTNTWVISQNCISNTCEGVASFNSKKSETFNPINEGLKIKFGTGIIKGVLGI DNITIGNDIIKQQIFGLVNEENSNIFNVSYGACTHRPRKGSSI
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TA05625 | T | 257 | 0.672 | 0.051 | TA05625 | T | 250 | 0.657 | 0.068 | TA05625 | T | 252 | 0.655 | 0.075 | TA05625 | T | 253 | 0.646 | 0.103 | TA05625 | T | 259 | 0.639 | 0.066 | TA05625 | T | 260 | 0.599 | 0.080 | TA05625 | T | 262 | 0.594 | 0.605 | TA05625 | T | 176 | 0.591 | 0.045 | TA05625 | T | 246 | 0.584 | 0.021 | TA05625 | S | 255 | 0.583 | 0.053 | TA05625 | T | 173 | 0.576 | 0.102 | TA05625 | T | 232 | 0.576 | 0.046 | TA05625 | T | 263 | 0.565 | 0.316 | TA05625 | T | 231 | 0.564 | 0.027 | TA05625 | T | 242 | 0.564 | 0.047 | TA05625 | T | 229 | 0.557 | 0.076 | TA05625 | T | 236 | 0.553 | 0.022 | TA05625 | T | 179 | 0.549 | 0.298 | TA05625 | T | 170 | 0.548 | 0.035 | TA05625 | T | 183 | 0.541 | 0.034 | TA05625 | T | 237 | 0.537 | 0.039 | TA05625 | T | 244 | 0.533 | 0.392 | TA05625 | T | 224 | 0.507 | 0.056 | TA05625 | T | 227 | 0.502 | 0.076 |
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TA05625 | T | 257 | 0.672 | 0.051 | TA05625 | T | 250 | 0.657 | 0.068 | TA05625 | T | 252 | 0.655 | 0.075 | TA05625 | T | 253 | 0.646 | 0.103 | TA05625 | T | 259 | 0.639 | 0.066 | TA05625 | T | 260 | 0.599 | 0.080 | TA05625 | T | 262 | 0.594 | 0.605 | TA05625 | T | 176 | 0.591 | 0.045 | TA05625 | T | 246 | 0.584 | 0.021 | TA05625 | S | 255 | 0.583 | 0.053 | TA05625 | T | 173 | 0.576 | 0.102 | TA05625 | T | 232 | 0.576 | 0.046 | TA05625 | T | 263 | 0.565 | 0.316 | TA05625 | T | 231 | 0.564 | 0.027 | TA05625 | T | 242 | 0.564 | 0.047 | TA05625 | T | 229 | 0.557 | 0.076 | TA05625 | T | 236 | 0.553 | 0.022 | TA05625 | T | 179 | 0.549 | 0.298 | TA05625 | T | 170 | 0.548 | 0.035 | TA05625 | T | 183 | 0.541 | 0.034 | TA05625 | T | 237 | 0.537 | 0.039 | TA05625 | T | 244 | 0.533 | 0.392 | TA05625 | T | 224 | 0.507 | 0.056 | TA05625 | T | 227 | 0.502 | 0.076 |
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| TA05625 | 178 S | NSTNSTKDP | 0.998 | unsp |