

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
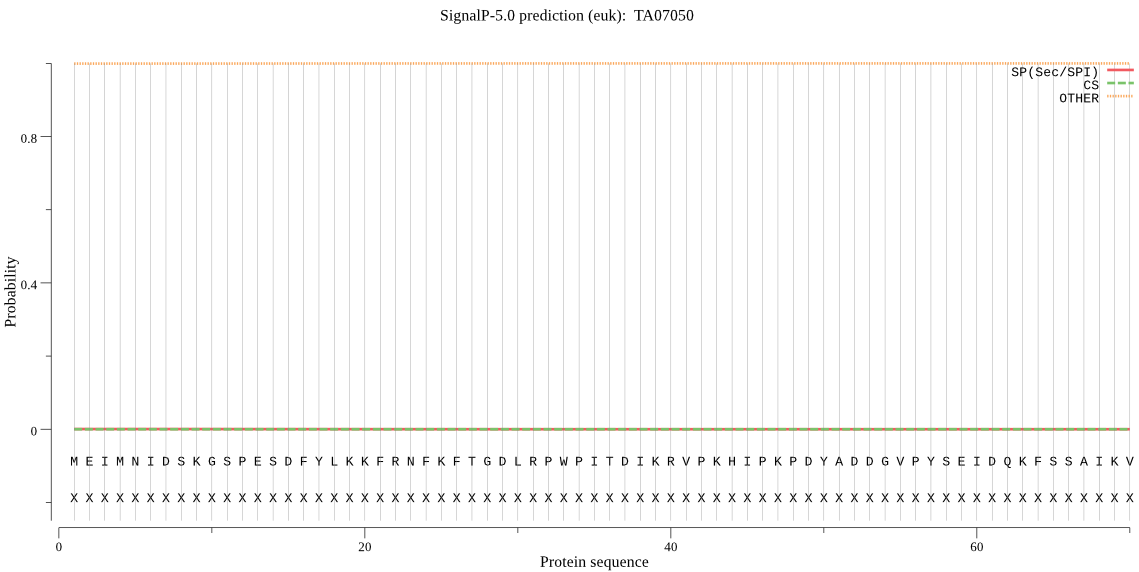
| TA07050 | OTHER | 0.999874 | 0.000052 | 0.000074 |
MEIMNIDSKGSPESDFYLKKFRNFKFTGDLRPWPITDIKRVPKHIPKPDYADDGVPYSEI DQKFSSAIKVHDPQTIKKIRRACLLGRKALDLANTLIKPGITTDEIDTKVHEFIVSHNGY PSPLNYYNFPKSICTSVNEVVCHGIPDLRPLEEGDIVNVDISVYLNGVHGDLNETFYVGE VDDDSMRLTEGTYASLMEAIKQCKPGMYYREIGNIINDVADKFGYFFNLILPRLSVIRSY CGHGIGTEFHCCPNIPHYRKNKAIGILRPNQVWSLNMSLGTFRDVKWPDKWTVVTTDGKR SAQFEHTLLVTNTGVEVLTKRLESSPPLGFDTTKF
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|
| TA07050 | T | 332 | 0.565 | 0.056 | TA07050 | T | 333 | 0.558 | 0.023 |
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|
| TA07050 | T | 332 | 0.565 | 0.056 | TA07050 | T | 333 | 0.558 | 0.023 |
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TA07050 | 58 S | GVPYSEIDQ | 0.995 | unsp | TA07050 | 58 S | GVPYSEIDQ | 0.995 | unsp | TA07050 | 58 S | GVPYSEIDQ | 0.995 | unsp | TA07050 | 235 S | LPRLSVIRS | 0.995 | unsp | TA07050 | 11 S | DSKGSPESD | 0.997 | unsp | TA07050 | 50 Y | PKPDYADDG | 0.99 | unsp |