-
-
-
Computed_GO_Function_IDs: GO:0008233
-
-
Computed_GO_Process_IDs: GO:0006465
-
-
Curated_GO_Component_IDs:
-
Curated_GO_Components:
-
Curated_GO_Function_IDs:
-
Curated_GO_Functions:
-
Curated_GO_Processes:
No Results
-
Fasta :-
>TA08609
MMNFLSRSYSVLHAGAMIGLIALFLNYYTGVNLRTNKHMIGDVKHIKTHELTLDKSKVDR
ALIELNMRYDLRGAFDWSTHMIFIYVTANYITNRHERSEVIIFDKIINNKSEAYQPSINI
FAKYFLYDFGRSLRNRDISLKFFYELVPIGGFIKQYQLSHNKFTMPKQYTQSS
- Download Fasta
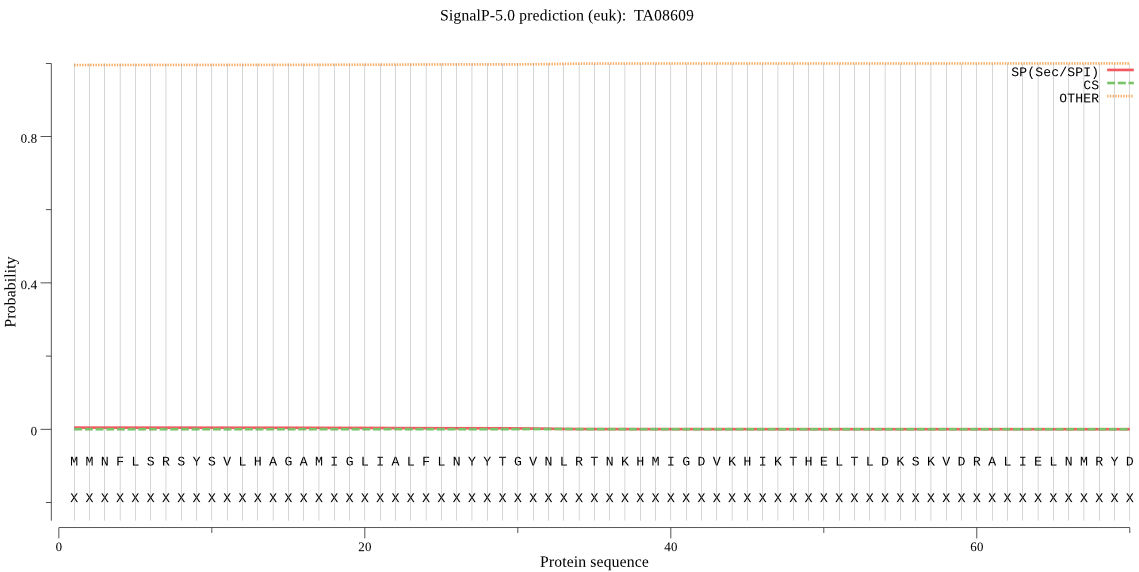
-
MitoProt II - v1.101
File : /home/rajan/sadaf/4480_mitoprot/test/TA08609.fa
Sequence name : TA08609
Sequence length : 173
VALUES OF COMPUTED PARAMETERS
Coef20 : 4.313
CoefTot : -0.837
ChDiff : 8
ZoneTo : 41
KR : 3
DE : 0
CleavSite : 0
HYDROPHOBIC SCALE USED
GES KD GVH1 ECS
H17 : 1.812 1.906 0.244 0.744
MesoH : -1.127 0.086 -0.580 0.117
MuHd_075 : 17.583 13.311 5.819 4.429
MuHd_095 : 21.422 12.913 4.862 4.763
MuHd_100 : 19.816 14.770 5.687 4.962
MuHd_105 : 23.343 20.654 7.538 6.422
Hmax_075 : 12.100 9.800 2.156 4.880
Hmax_095 : 6.500 5.162 -1.559 3.019
Hmax_100 : 8.200 16.000 2.943 5.740
Hmax_105 : 13.300 12.600 1.660 4.900
CLASS NOT-MITO MITO(/CHLORO)
DFM : 0.4366 0.5634
DFMC : 0.5010 0.4990
-
Fasta :-
>TA08609
ATGATGAATTTTTTGTCTAGAAGTTATTCAGTATTACATGCTGGTGCTATGATAGGATTA
ATAGCATTATTTTTAAATTATTATACTGGTGTTAATTTACGTACTAATAAACATATGATT
GGTGATGTTAAACATATCAAAACACATGAACTTACACTTGATAAATCTAAAGTTGATAGA
GCATTAATAGAATTGAATATGAGATATGATTTAAGAGGAGCATTTGATTGGAGTACACAT
ATGATATTTATATATGTAACAGCAAATTATATAACAAATCGTCATGAAAGAAGTGAAGTA
ATAATATTTGATAAAATAATAAATAATAAATCAGAAGCTTATCAACCATCAATTAATATT
TTTGCTAAATATTTTCTTTACGATTTTGGTAGATCACTTAGAAATCGTGATATTAGTCTT
AAATTCTTTTATGAATTAGTACCAATTGGTGGATTCATTAAACAATATCAACTCTCACAT
AATAAATTCACTATGCCAAAACAATATACACAATCATCTTAA
- Download Fasta
|
ID | Site | Peptide | Score | Method |
|
TA08609 | 139 S | NRDISLKFF | 0.995 | unsp |


