

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
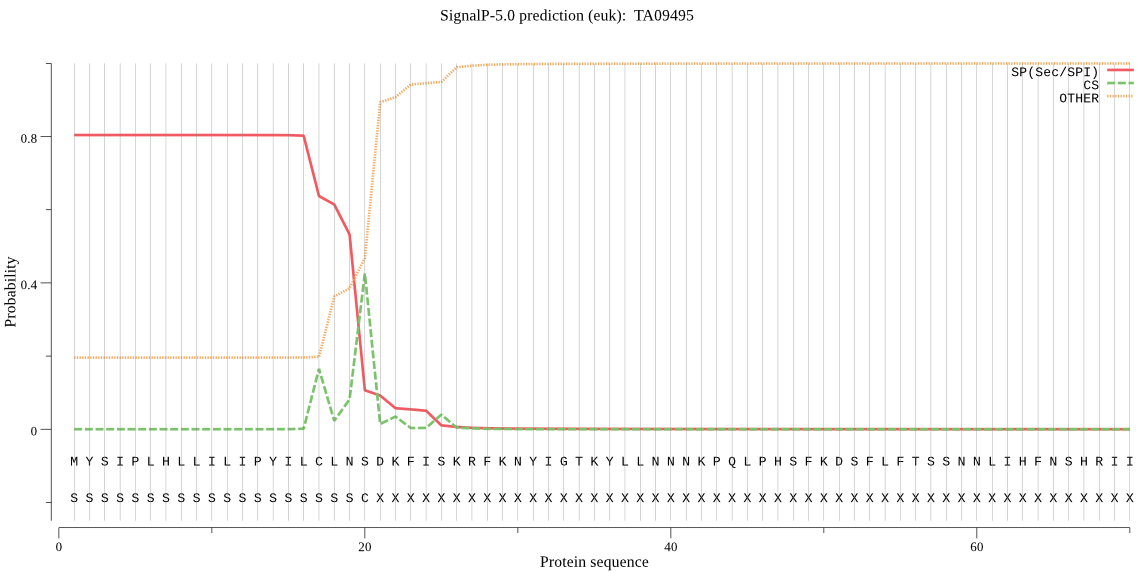
| TA09495 | SP | 0.032062 | 0.967437 | 0.000501 | CS pos: 17-18. ILC-LN. Pr: 0.5474 |
MYSIPLHLLILIPYILCLNSDKFISKRFKNYIGTKYLLNNNKPQLPHSFKDSFLFTSSNN LIHFNSHRIITYIFSHKHILNNERFIRSIPPLGFSPTCSISYLNQSNRTHEIIQKLKEEA PNASLFDIWNNFCYSGWFKYSTILTGRIRKGVLSGKQFPSKNIMRPNYFKTGKPIYVDYP YCNRDSKDIRGTIKNDKDVAGIKKACRIAREILDYVSSIVVEGIFTDDIDRFVHKLCGIK KVFPATLNYHGFPKSVCTSINEVACHGIPDNNLLCAGDLLKVDLTVYSDGYFGDVCETYM VMPMTKEYNKKLLQTNYMGRSERNKLIYTSVRHNNLQGCEISIIKQTSFINGFNNAIKRF DLENTFNLLYLILKGEDKRFNFSQDKNMSKLAKKLNEEFKAVYENPEETYSGRVFGMPSS ELPGYIPHHKLGKVFATVENPQIVVDLLDQNDELRFKKPYTFSPSFDSDLELMKICHEAL MKAISICKPGTKIKMIGKTIEKYLKKNKCISLSNLCGHGIGRNFHENPIISHEENDSEVL MEPGMVFTIEPIVTRSGLNSFMMWPDGWTIATLDGSKTAQFEHTVLITKDGHEILTKKIT SSPQFIWERELEVVY
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TA09495 | 531 S | NPIISHEEN | 0.995 | unsp | TA09495 | 531 S | NPIISHEEN | 0.995 | unsp | TA09495 | 531 S | NPIISHEEN | 0.995 | unsp | TA09495 | 48 S | QLPHSFKDS | 0.998 | unsp | TA09495 | 154 S | KGVLSGKQF | 0.992 | unsp |