

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
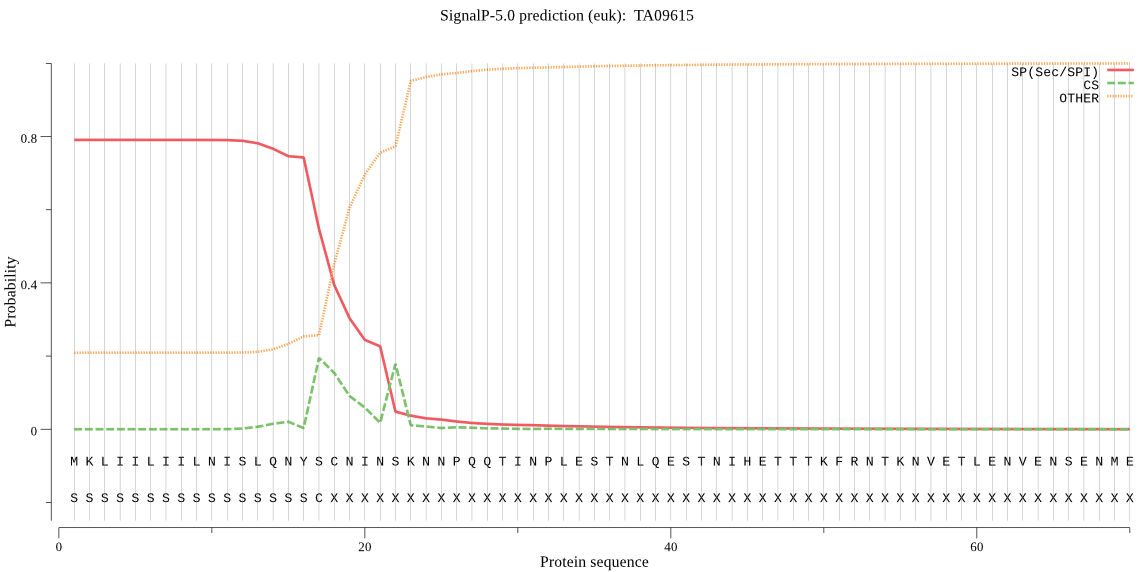
| TA09615 | SP | 0.064038 | 0.935452 | 0.000511 | CS pos: 18-19. YSC-NI. Pr: 0.3243 |
MKLIILIILNISLQNYSCNINSKNNPQQTINPLESTNLQESTNIHETTTKFRNTKNVETL ENVENSENMEQSEKLDEIEFDEKLVLNILKRRPKGFEVIRQNYYGIDILVFIPYGIITEI IDSNGIIWKNYNNIKYIKVTTFKHNSKLLHIQFFNSPVTVLGQTDSNGPKVSSSKGISST TGTVGASTVTEGKGANDTFSTPGKGANSTLMECTPGKGANFTLMECTSGKGANSTATECT SRKGANSMPMECTPGKGANSMPMECTIGNSTEGSKGAIGTRFESQSSNTTGREPDTVTEV TEIYYHRNGRVWNEIDEENFYNIFNIIRMESSCLGKYTIYLNDKNSHRNVIWNHSDERYA TYAPASGIRINKLCDYNWGPICLKSIWRTKKNEYLLYTSVFNKIKPKLVYIILSNGIDIT EEYYFKYNSKWYQINRNTFFNKLYTYFPVTVTGPPESSTQDSTVIPSTSTNGGTIGASTV TEENSTTKIAAPKVGTTVLTEPLGTSTTNSTGKGANDTFGSPGKGANSTLMECTSEKNST EIAAPKVGIGPFGTRFETQFSNTNTIRGPDTVMEKLNKLMEIEIDFENINKEKLYTSTFG INRTIQIIYPRPGIKIISVRNGKSIIWKSNFNLYCDYAYLIKTPELPILCYLLLSNNFTR KINYFIYLNYWININKSQFFYFITHNISTSQILKIISNNTKNTNNIEDSIEDTENEEDSI EDSRDKDINVESTVKEDISVESKVNTDSILESTVNYENEEDSIESEDELFTVKEIPIESA KEGEITIESENENTEVNSTKDTTGKGANDTFGTSGKGANSMGMECTTGKGANSTAMECTS EKNSNEIAVVTKTGESSTFSEESKDTMDTNTNTKETPIRAEGEDTNTKEAPIRAGTRDTK GVDEETGTVGASTVPEIETSNFINKKGLKIRIYRSRIKESKGIIILTHGIGLHFIEVSMR NNLEWNYENFGFITHPFLICPNLNYYNLNNLNINRFKHIFQHNFFSSNISTNLSPNILST TNISSTTNKSENISENSKKSNKSENLTNLSTNKSNISEKSNLLPNYEYKNSLMELLNNLG YDVYGMDLQSNGFSESYDSIRSHIHNTNDFLDDLLQFIFIIINNQFYSNNNNLNNNKFNL NKFENLNNFVNLINWKKKKRIILFGFSFGANLSIRIIQYYYQSIYKMKKKYKKFGLNKRF GLNKKIKEKIGKFEENKKIKEKNEELDNIFVEDLLVDGVISLSGMLNMDNHLGTRFIKLK ILLIKFLALICPRNINKFKSLPVFNQSYQYSILLNVRSYGACYSSSQSTTKPLVILITIV LK
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TA09615 | T | 463 | 0.626 | 0.022 | TA09615 | T | 468 | 0.573 | 0.194 | TA09615 | T | 459 | 0.568 | 0.176 | TA09615 | T | 470 | 0.535 | 0.072 |
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TA09615 | T | 463 | 0.626 | 0.022 | TA09615 | T | 468 | 0.573 | 0.194 | TA09615 | T | 459 | 0.568 | 0.176 | TA09615 | T | 470 | 0.535 | 0.072 |
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TA09615 | 762 S | NEEDSIESE | 0.995 | unsp | TA09615 | 762 S | NEEDSIESE | 0.995 | unsp | TA09615 | 762 S | NEEDSIESE | 0.995 | unsp | TA09615 | 779 S | IPIESAKEG | 0.994 | unsp | TA09615 | 798 S | TEVNSTKDT | 0.997 | unsp | TA09615 | 860 S | SSTFSEESK | 0.992 | unsp | TA09615 | 1099 S | ESYDSIRSH | 0.992 | unsp | TA09615 | 709 S | NIEDSIEDT | 0.997 | unsp | TA09615 | 719 S | NEEDSIEDS | 0.998 | unsp |