

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
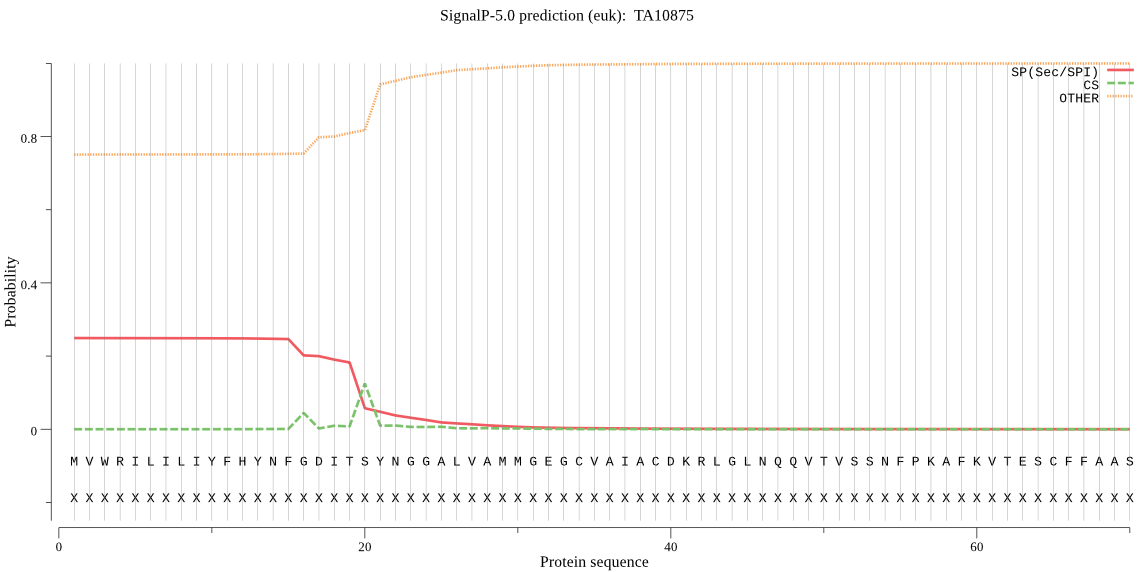
| TA10875 | SP | 0.394313 | 0.605292 | 0.000395 | CS pos: 20-21. ITS-YN. Pr: 0.6692 |
MVWRILILIYFHYNFGDITSYNGGALVAMMGEGCVAIACDKRLGLNQQVTVSSNFPKAFK VTESCFFAASGLATDVQTLKDEIMFKVNMYKLRGDKEMSVKTLSNMVGSMLYSRRFGPWF VDSVIAGLDTDSSPYITCFDLVGAPCTPTDFVVAGTCSEQLYGVCEALFKPGMDPEQLFE TVSQCLMAGIDRDCLSGWGAEVHVITPDRVISRSLKTRMD
| ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|
| TA10875 | T | 217 | 0.522 | 0.031 |
| ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|
| TA10875 | T | 217 | 0.522 | 0.031 |