

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| TA11060 | OTHER | 0.999850 | 0.000150 | 0.000000 |
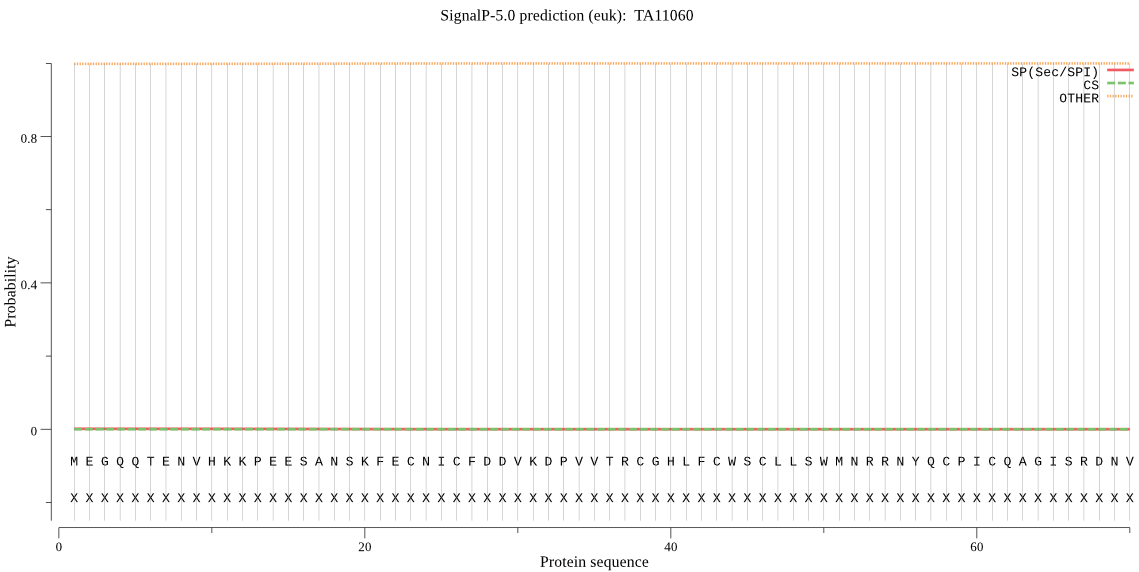
MEGQQTENVHKKPEESANSKFECNICFDDVKDPVVTRCGHLFCWSCLLSWMNRRNYQCPI CQAGISRDNVIPLYGHGQNQSDPRDKPEEPRPKAQRSTNNQRQNSFFRGYENRISVSFGS FPFSFIFPIAFGTSNTGSFFDLFRNDESSSNMTSEIFVKICSEQRRAYAYTGFLSLIGLL MIAYLILCI
| ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|
| TA11060 | T | 6 | 0.538 | 0.081 |
| ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|
| TA11060 | T | 6 | 0.538 | 0.081 |