

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
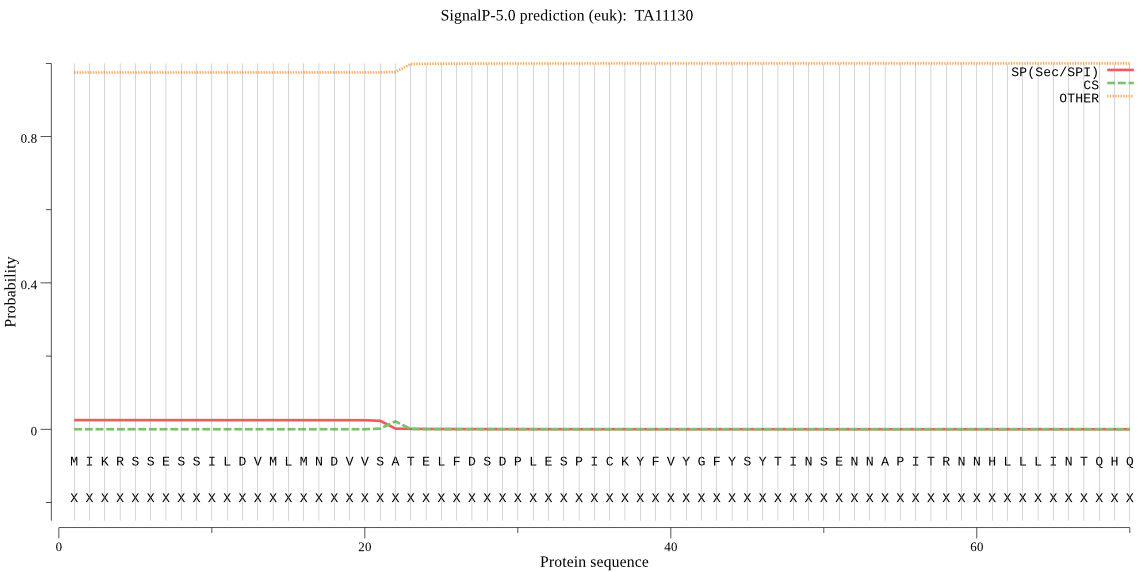
| TA11130 | OTHER | 0.998346 | 0.001636 | 0.000018 |
MIKRSSESSILDVMLMNDVVSATELFDSDPLESPICKYFVYGFYSYTINSENNAPITRNN HLLLINTQHQINFYPINQYNKDLNSHNSQRNPTKRLETRDGYMDSDDGKSYYDCVVESDE QHSESDVVIQNDDVIEADYDVIELKKNTLNLRKIYMPGKPSKTIAKNELQLIIESPVKVG TCEFNFCALLETGNKLTLFYYDSAELEYLNDSDTITLDHITIKDVFPTVKSVYNECTIST FSEIYHLLSSSLKRFKFRLKGFDEVENLFDVTLTNYKIINSILETVNYDGLCINKCYLDE KNVESFSSFSYLDDSIIDFFNQFTYKYLMDESQRKTWVILSTYFVRKIKQYKDPKEAYTN TWKWTRKFTRALPMNDFIFIPINLSEVHWSLVIIAYPKYAIRYHSIKSEKKASIIHLDSL GNHHLSHDIIDLLKNYLYQEYDNRCRIFKERGFEFDLDPDSWDYIAPSRGVPLQNNGYDC GIYLIEYIMYLTRNKNEFSTLIPKYFEPKNVNRTESGRYGKWFTQVQIHNRRLSMKQVLK FMRDNPNWSTDQNKIQYVIDQMTQQY
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TA11130 | 123 S | DEQHSESDV | 0.997 | unsp | TA11130 | 123 S | DEQHSESDV | 0.997 | unsp | TA11130 | 123 S | DEQHSESDV | 0.997 | unsp | TA11130 | 315 S | YLDDSIIDF | 0.994 | unsp | TA11130 | 405 S | IRYHSIKSE | 0.997 | unsp | TA11130 | 516 S | NRTESGRYG | 0.99 | unsp | TA11130 | 534 S | NRRLSMKQV | 0.996 | unsp | TA11130 | 6 S | IKRSSESSI | 0.995 | unsp | TA11130 | 110 S | DDGKSYYDC | 0.996 | unsp |