

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
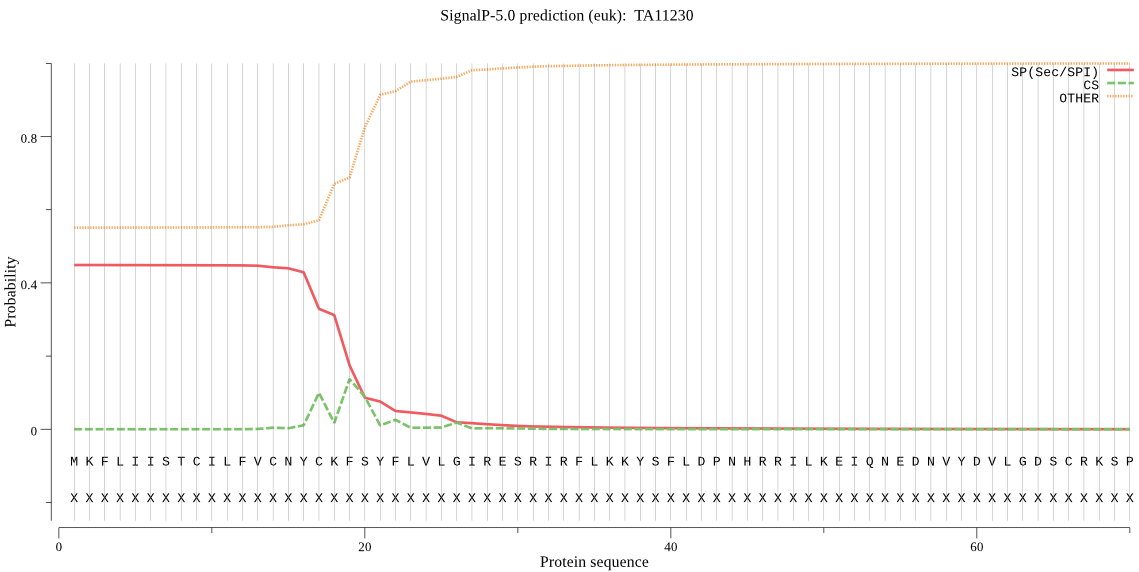
| TA11230 | SP | 0.346438 | 0.652397 | 0.001165 | CS pos: 17-18. NYC-KF. Pr: 0.4773 |
MKFLIISTCILFVCNYCKFSYFLVLGIRESRIRFLKKYSFLDPNHRRILKEIQNEDNVYD VLGDSCRKSPPLYSTYSEYLRDNQVIQIEQEPKSLKNILSIETSFDDTCIAVVRSDGKIL SDKKLSQEEVVKEYGGIKPVCAKLEHIKKIESLTDKVIEESGLKIQDIDEIAVTRGPGTE LCLRVGYNYAKELSEKYKIPLVSENHIAGHCLSPLIDEHQFKYTVEGTPIKSNDLKFPYL CLLLSGGHSQIYLVENPSKFHLMCETQDEFVGNVLDKCAKLLGLDLSKGGGAELEKIADE VSDSKYKLTIPNKYNHYMEFCFSGVQSQLGLKTEQLVKSHNVEDAKRLPRKILSELAYGL QSTVFEGILIQLEMSLNAVETLFPINQLALVGGVASNDKLKKMILDLFYLRDESVRFSEQ EMFLTRTKNMVKRYCKIGGNYLTSGNPYYKFFQDFVKNVTNLEDFIYYLCNPELYPEVLV PGYIIKIGKEHKRELYKSMLCTYFDLIGLKNVFKIKHKIGKYFKSFELIKPSKRILNLIE NDSKFTLPLRRHGDEISDRRWDLYTTSKKYCTDNAVMIGFSLIQKNRMGIKEINSPEKIN GKDVAPRWDLGTRKNWELLSDICKLDSLIK
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TA11230 | 152 S | KKIESLTDK | 0.993 | unsp | TA11230 | 152 S | KKIESLTDK | 0.993 | unsp | TA11230 | 152 S | KKIESLTDK | 0.993 | unsp | TA11230 | 414 S | LRDESVRFS | 0.995 | unsp | TA11230 | 418 S | SVRFSEQEM | 0.996 | unsp | TA11230 | 94 S | QEPKSLKNI | 0.994 | unsp | TA11230 | 126 S | DKKLSQEEV | 0.997 | unsp |