

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| TA11350 | OTHER | 0.999911 | 0.000047 | 0.000042 |
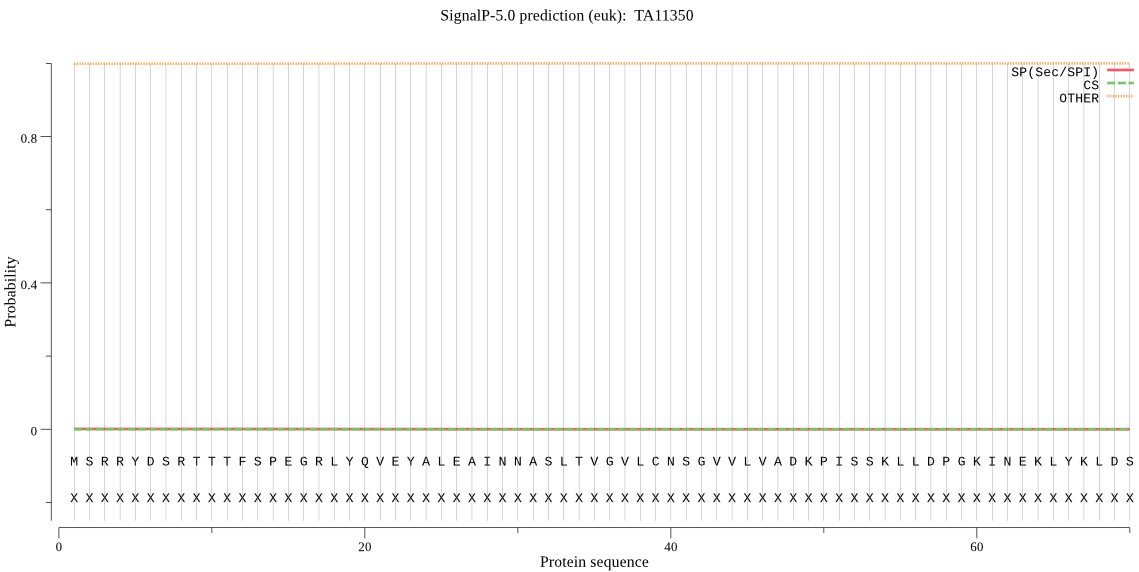
MSRRYDSRTTTFSPEGRLYQVEYALEAINNASLTVGVLCNSGVVLVADKPISSKLLDPGK INEKLYKLDSHIFCAVAGLTADANVLINMCKLYAQRHRYSYGEPQSIEQHIIQICDLKQS YTQFGGLRPFGVSFLFGGWDENLGFQLYHTDPSGNYSGWKATAIGMNSQSAQSILKQEWK EDLTLEQAIKLAIRVLTKAMDSSTPQADKIEVGVLSKGETGEFEPFVEMLNNERVDTLLK TIAQEDTNKSTNTQ
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TA11350 | T | 253 | 0.524 | 0.020 | TA11350 | S | 2 | 0.514 | 0.085 | TA11350 | T | 251 | 0.504 | 0.018 |
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TA11350 | T | 253 | 0.524 | 0.020 | TA11350 | S | 2 | 0.514 | 0.085 | TA11350 | T | 251 | 0.504 | 0.018 |
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TA11350 | 100 S | RHRYSYGEP | 0.993 | unsp | TA11350 | 100 S | RHRYSYGEP | 0.993 | unsp | TA11350 | 100 S | RHRYSYGEP | 0.993 | unsp | TA11350 | 7 S | RRYDSRTTT | 0.995 | unsp | TA11350 | 13 S | TTTFSPEGR | 0.99 | unsp |