

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
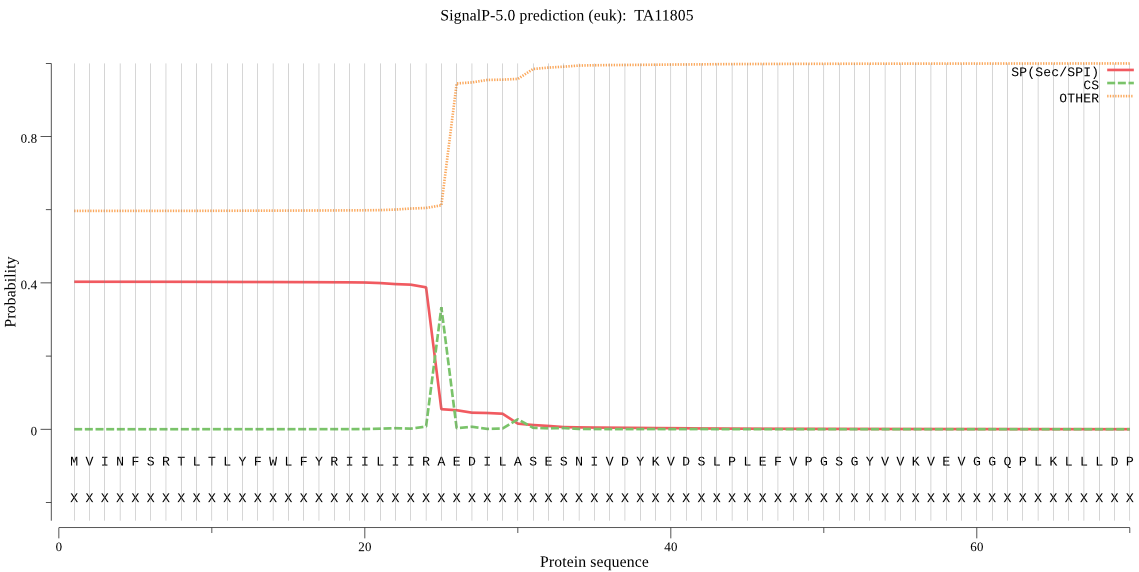
| TA11805 | SP | 0.211742 | 0.787550 | 0.000708 | CS pos: 25-26. IRA-ED. Pr: 0.8947 |
MVINFSRTLTLYFWLFYRIILIIRAEDILASESNIVDYKVDSLPLEFVPGSGYVVKVEVG GQPLKLLLDPNVCGIILFENTDRICSKDDKGSCYDPYKSKTASWCVNTAVCVPGKFNYQC KETPSPSKIKELTVDSDIIKIYSIEGLESLKIAVDHKKSPYILDKVPVKLGRSLDRYDRK IFTNVDGIFGISVTRDYRGFFVLDINPVQNVRFPSKLFLGTDRVSEDEIVWSEKRQTGGI FTNSLIQFTIYDLKMCNTKIFGRTSSNWEAAIDLTTPYLILPKNFWMTMMSYLPVDKSCF DEGLSPRLCKLTVGNRLFPIIEFKLSESYYLNFEKVETPSITIPLENLIYDDGDSKTLLI IPDEFSDRPSYTLNPTIKFGYKVLESLNVVVDSDGYRVGLISKNQLVGSFSKCSEVPQCF GDQVYEPALNICLNPICSIWLMKRLNPEKGICETSFVAKVVITTVICALVVAELYCNFAR KHILRITSRLCR
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| TA11805 | 92 S | DDKGSCYDP | 0.992 | unsp | TA11805 | 225 S | TDRVSEDEI | 0.997 | unsp |