

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
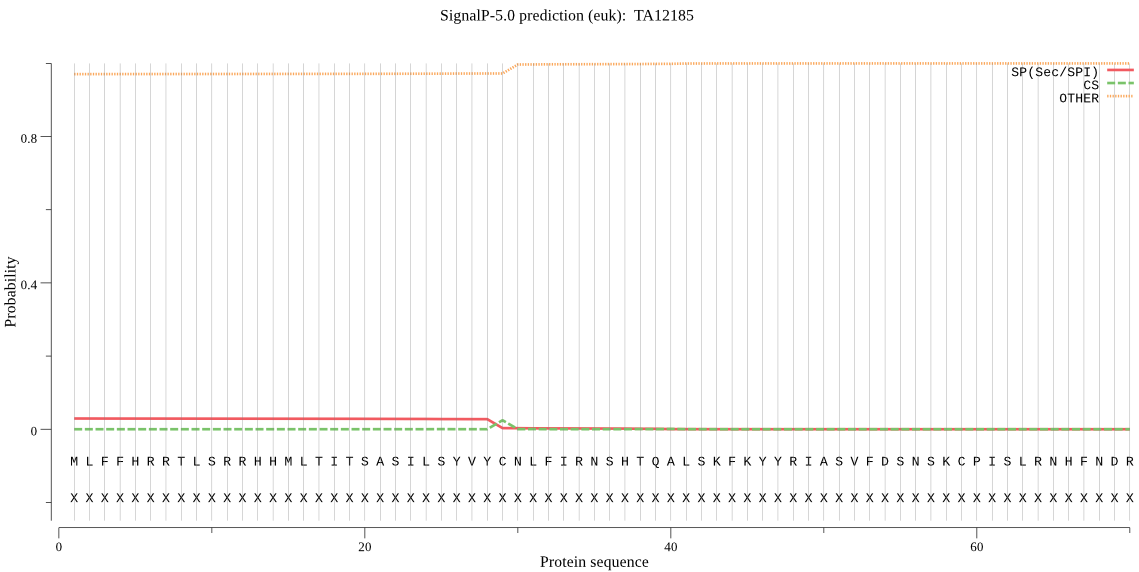
| TA12185 | mTP | 0.166605 | 0.001693 | 0.831702 | CS pos: 79-80. RLY-SS. Pr: 0.7675 |
MLFFHRRTLSRRHHMLTITSASILSYVYCNLFIRNSHTQALSKFKYYRIASVFDSNSKCP ISLRNHFNDRYYLFIRRLYSSSPANNVEDVLYKVFSPDSFSETTFRHSILNMKFPDQADE PEWVQESKSFNHPSFEKIGSVYLPDIGVVASNYKHKLSGLSVFSLKSHIDSGKEMCFDLI VPSPPLNSKGSPHVLEHSVLSGTPKYPMKDPFSLLVQGGFNSFLNAMTYKDRTSYLFAST NEKSFYQTGDVYMDSFFRPNITKDKTIFEQECWHYKVTDGTSDKSDADVPLHGRMIGYSG VVYSEMKNRFSDSSCLFYNLIYQNLFSNSYKYVSGGDPSDIVDLTHQELVNFYKLYYGPK TATLYFYGPYDVQNRLDFVDKYLTTYNIGIEKDPNNENLVHNASLLSSDSNLPYEEYKSK PKHVSSEYSSVDPTEDELMISWLLDPLYNGSMDKFKIDPVDNVGFQVLQYLLLGTPESVL YKGLIDSGLGKKVLVHGFLSGYKQSLFSFGLKGVDNTKHNSKDEIVRKFEEVVFGILNKI TEEGFKRDAIDSGLNLVEFEMRELNSGSYPKGLMLIDLIQSQLQYGKDPFGLLRFDSLMK ELRSRIFSDNPSNYFINLIVKHMLNNNTRVTVHLQAVEASKYEKEFNKKIADQLRERLSH LSKEQVDEMEEYYKKFKAEREDMDINDGSESLKTLELSDISREQETIPTKFYKLSSDGLS ESNALYNDGKTFTVLTHPIDSHGVLYMDYALSLDSLTVDDLCYLNLFSSMLKESGTDKLT PEELTYKIDKNLGGLSLSTYFTTETNNKTYDDPEDGLGYLIVRAKCLKHKVNEMLEVVNE VLLNADFSNSKKGLEILKRALSMYQANVSSKGNEFALRRMCAKFSVSDYADELVNGYSQL VFLRDTLVPLAEKDWSKVESKLNEMRVKLLSMKNLTVNLGGDSELLDSVLDDSTTFYSKL SSTFKYGSKTSDKVWVKEVLDKKLMDSVDKNELIVVPSRVNFVGMGGKLFDKNDEVLGSN SLAVHYLSRKHLFTFVRMSLGAYSVYSYLLNTGHIIFMSYADPNFEKTLEVYRNLASVMK EAYEKIEDSELLRQKIGKISGLDKPLHVENKTEVALRRALRKESDEFRQKFREDVIDSTK ECFNRLYKQMTDQKEWNNVSAVVNSNTSDEAPSDYKRLQIN
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TA12185 | 285 S | TSDKSDADV | 0.992 | unsp | TA12185 | 285 S | TSDKSDADV | 0.992 | unsp | TA12185 | 285 S | TSDKSDADV | 0.992 | unsp | TA12185 | 334 S | YKYVSGGDP | 0.997 | unsp | TA12185 | 916 S | EKDWSKVES | 0.99 | unsp | TA12185 | 931 S | VKLLSMKNL | 0.993 | unsp | TA12185 | 108 S | TFRHSILNM | 0.993 | unsp | TA12185 | 171 S | SHIDSGKEM | 0.997 | unsp |