

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
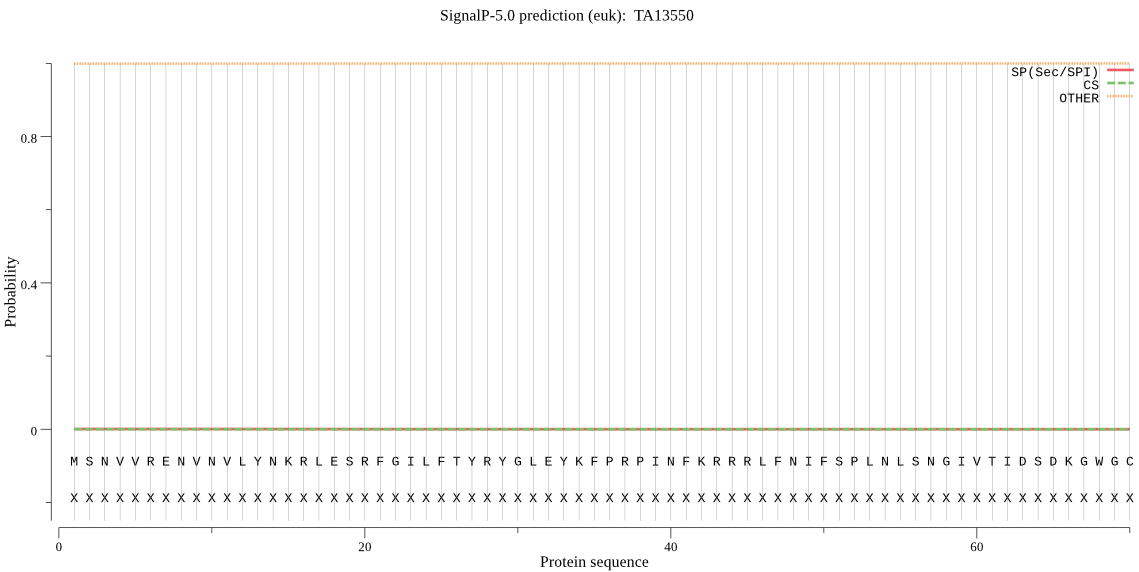
| TA13550 | OTHER | 0.998900 | 0.000081 | 0.001018 |
MSNVVRENVNVLYNKRLESRFGILFTYRYGLEYKFPRPINFKRRRLFNIFSPLNLSNGIV TIDSDKGWGCVLRSTQMAISQALLNLVLGPEFSVEQLEIRNRTPRNRKIDQSLLNIDTFE KLLNGLLDLDGVSAVSVILAQFYDDLNAVFSIYNFVIADYVLKTCTKFLHFGPTSAALCA SKIINDLNLPINSIAFPDGVFHISDVREILEEKRNLLVWVSNKKKLDRIERECVRSMFRL SQFNGIIGGNLFNKSYYIFGTTNKRLYYNDPHLYCKKAFRSLEYVDIFRDFTSRRVKSMN WRYFNASFTLLFLFKDRDDFQDFLENTFERKSIVESFPFEFITYGDLTFN
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| TA13550 | 332 S | FERKSIVES | 0.997 | unsp |