

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
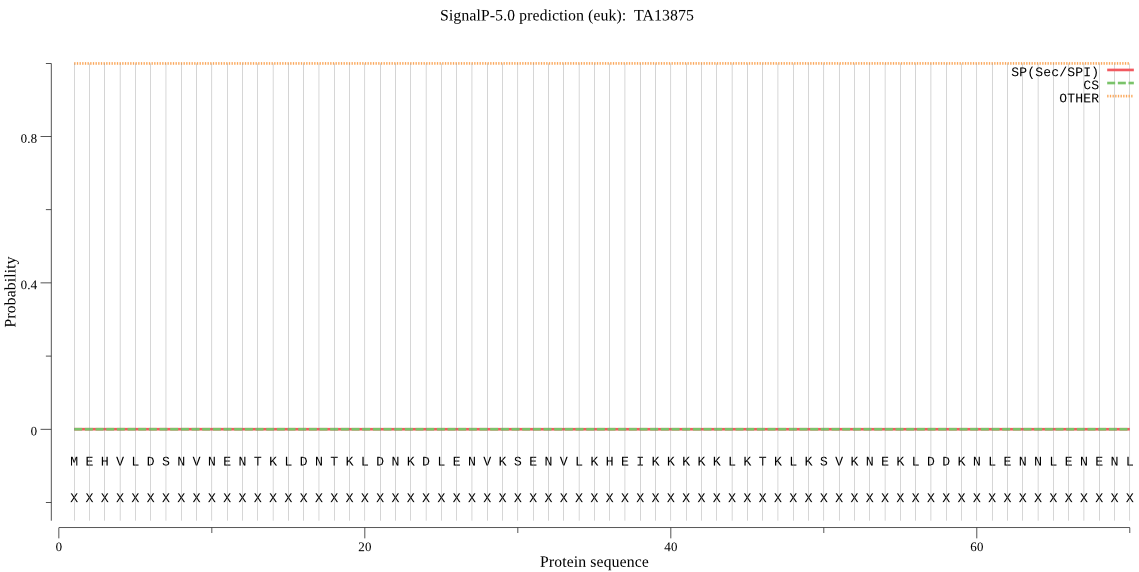
| TA13875 | OTHER | 0.999997 | 0.000001 | 0.000002 |
MEHVLDSNVNENTKLDNTKLDNKDLENVKSENVLKHEIKKKKKLKTKLKSVKNEKLDDKN LENNLENENLENEKPKIKKNAKLIKKTQLTNTNTTGTNTNTKGKGANDTFDTPGKGANFT ATECTTKDTKGVGEGTGTVGPSTVTEDKNISREKVAKLKKKFETKKNQSTYNKVINLFKQ HNLLTEQQNDEKNQLDDKKSSDDKKILNNENNSLNDKNSLDNVIINIESNTNKQNNTTNT TTGTTTNTTTATTTVGMNKIKGVKKITTVFGGYPVLTLTSSVILFVFLFQIILNKISFNG RCISKVLYPNDSKAYHTMFGYGACEYNLQQKIEEIGYIGTKANDKGWPSTLVSDENVVES TSTFDSPNYRGFTLFGGMNTNYIRNYKEYFRLFWSMFMHSGVLHLIFNLIAQTQILWIIE PDWGFFRSLSTFILSGFMGNLFAGVFEPSFTVVGSSGCMFGLIAALIPYCVENWSLLRSP IYIFAFAIIITIISFTFFNNTVSIYSHLGGWVGGLLWGFITLRSTFKLKDCAIKESLLLS FFKDKLNPDDKNKLKESFLIKQRKCINEIRIERRRKKFKQRIKNLIKYPIDYFKKGPYDI LIRIIPLILFVILIIILCSNLLIESIYNRMFLLNESSNLSQCCYTNAKSEIIFNRKVFWC FENYQTAINFCKI
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TA13875 | T | 242 | 0.523 | 0.038 | TA13875 | T | 240 | 0.509 | 0.020 | TA13875 | T | 241 | 0.505 | 0.082 |
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TA13875 | T | 242 | 0.523 | 0.038 | TA13875 | T | 240 | 0.509 | 0.020 | TA13875 | T | 241 | 0.505 | 0.082 |
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TA13875 | 362 S | VESTSTFDS | 0.993 | unsp | TA13875 | 362 S | VESTSTFDS | 0.993 | unsp | TA13875 | 362 S | VESTSTFDS | 0.993 | unsp | TA13875 | 50 S | TKLKSVKNE | 0.997 | unsp | TA13875 | 200 S | DDKKSSDDK | 0.992 | unsp |