

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| TA14360 | OTHER | 0.999873 | 0.000126 | 0.000001 |
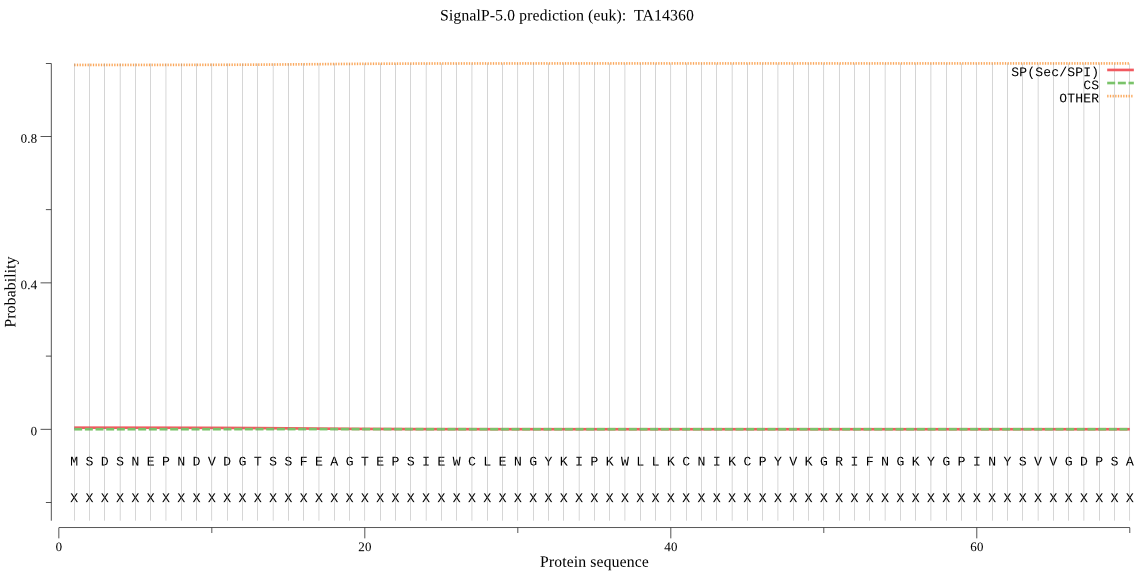
MSDSNEPNDVDGTSSFEAGTEPSIEWCLENGYKIPKWLLKCNIKCPYVKGRIFNGKYGPI NYSVVGDPSASLVLTFHGYNATHTTFSIYQSVLSKNGFRVISFDLYGHGLSGYPKYKVFG NTFSSKYYVDQADEVIDHLGYTGRKLSVIGMSMGACIAASYCETHPDLVEKIILISPAGL IPKKPMRVVFLKYIQCCIPCTPLCVSKCCFSGRVTTPKSKSNRESPESSQESLETGECVS VDEVSGESTELNPMLNRMLWTLFVTRRAISNLLGIVNRMPLWSSRELYRRVGGMGKLTLI LFGDSDTLTPPECADELSRLFTNSHTIIFSNSDHLLSFKKPLQVVSTCLSFLGIPNQENA QNYTRWLPFDCNGVYIPKSRRELGHSDETELTHNVSAPSLGSLTNHALPDEPPKSSTTSP FKIFRDYLPPDRTIKQLSLTEEYPRKKVPLVVVKQFENS
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TA14360 | T | 404 | 0.589 | 0.052 | TA14360 | T | 235 | 0.509 | 0.075 | TA14360 | S | 2 | 0.503 | 0.100 |
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TA14360 | T | 404 | 0.589 | 0.052 | TA14360 | T | 235 | 0.509 | 0.075 | TA14360 | S | 2 | 0.503 | 0.100 |
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TA14360 | 225 S | SNRESPESS | 0.998 | unsp | TA14360 | 225 S | SNRESPESS | 0.998 | unsp | TA14360 | 225 S | SNRESPESS | 0.998 | unsp | TA14360 | 228 S | ESPESSQES | 0.995 | unsp | TA14360 | 229 S | SPESSQESL | 0.99 | unsp | TA14360 | 232 S | SSQESLETG | 0.993 | unsp | TA14360 | 245 S | VDEVSGEST | 0.995 | unsp | TA14360 | 438 S | IKQLSLTEE | 0.993 | unsp | TA14360 | 216 T | GRVTTPKSK | 0.994 | unsp | TA14360 | 221 S | PKSKSNRES | 0.998 | unsp |