

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
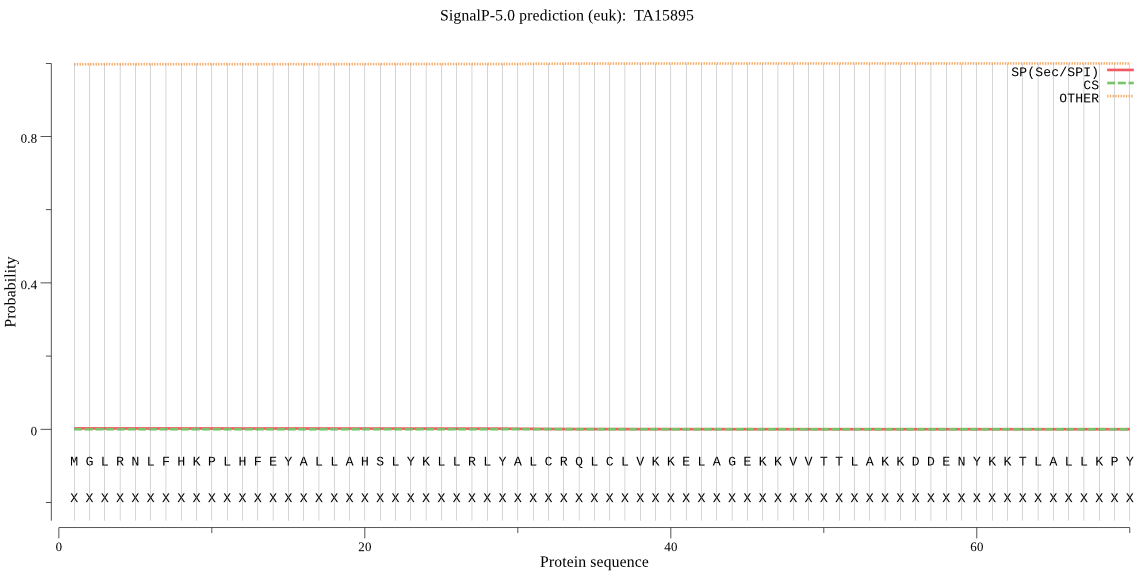
| TA15895 | OTHER | 0.937139 | 0.001919 | 0.060942 |
MGLRNLFHKPLHFEYALLAHSLYKLLRLYALCRQLCLVKKELAGEKKVVTTLAKKDDENY KKTLALLKPYLTSAAYQKTLEYSKDKLKFDMTFELVHWLVMFAFMFNNNVLKYWHLSGEL LRHKCHYSQVLVYFALRLGFSFLFRLPFRYYTAYRLEKKHGFKTKSRFVFLKQYLLCYAF YVLLLTGLASGLTWLNKFSKSNFRALAFLVFFKTALVFVVPLFLTLKHKLLPLSDPELRR EVDAMGKKLGLTSKNVHVVSANTAHTHGVSLSWGFCKFKHAYLNESYVTLGKPSAMALLS HVFGHFKHHHFFKSFLFDLTKGTVFLFLFDHFKGDTALFKSFGTHSVSALTMKLETVYMA YLCPLLLLVVVVKSVYSHFLEFEADRHAVRLGHSDELVNFWTTVYREKKWFFNVDPLYGR LFSERPSLFERVYAVYDATVASKPVSN
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| TA15895 | 427 S | SERPSLFER | 0.993 | unsp |