

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| TA18080 | SP | 0.172857 | 0.825951 | 0.001191 | CS pos: 22-23. IKC-FR. Pr: 0.9190 |
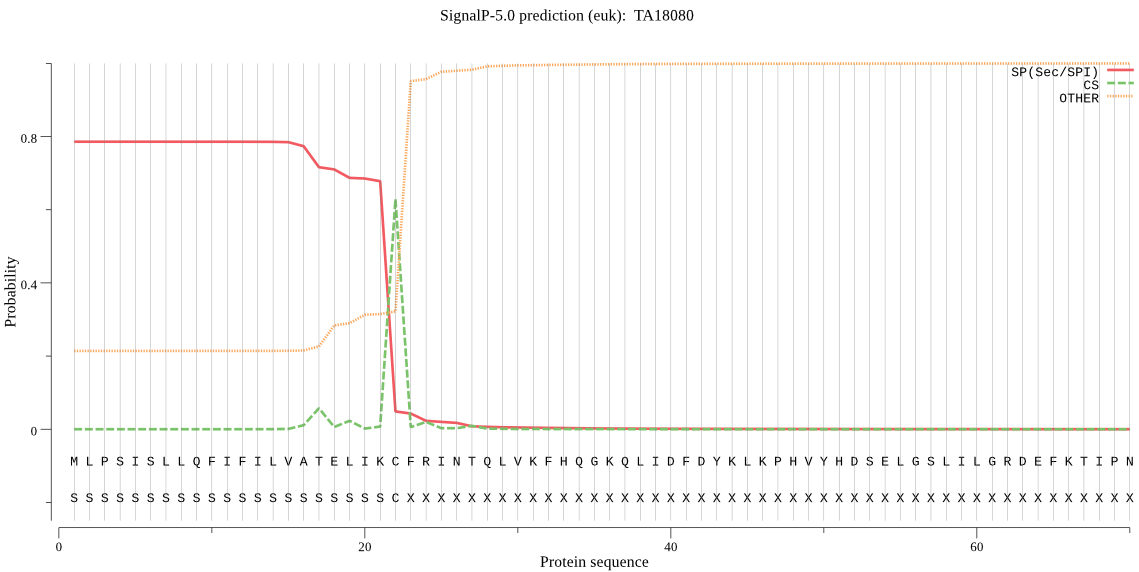
MLPSISLLQFIFILVATELIKCFRINTQLVKFHQGKQLIDFDYKLKPHVYHDSELGSLIL GRDEFKTIPNYLRYENLTNFITDKAELVQNLKRNLSRYRNLTVKTVEKFKRSSLANFRKC YSDLWEEIKFWYLKGSNFSFYDLFTLGSNPVYLLVKLNLALFIVDLFFKGELSKNFSVKG LNLFYMNEHYRIATSLFFHWDLRHLLSNVGSLLSVGNNVLKLFGPRNLLIIYFVSGITAN YFSYLYNYIYKNGIPANLLLLLRSIGKNFSNATKPIIMATNTIVDDILNKTRRIGFPMFF FDYIFTSATMSADFCLAKLAQFVTGITPRSLNDVDLKVKEICMTKSKNDFKSCGASSSIF GLYGALLTYYIKSGYINNYSVINNLFVSFVVPYLLRSFTEKLDHISHLVGFITGSILSTV LD