-
Computed_GO_Component_IDs:
-
Computed_GO_Components:
-
Computed_GO_Function_IDs:
-
Computed_GO_Functions:
-
Computed_GO_Process_IDs:
-
Computed_GO_Processes:
-
Curated_GO_Component_IDs:
-
Curated_GO_Components:
-
Curated_GO_Function_IDs:
-
Curated_GO_Functions:
-
Curated_GO_Processes:
No Results
-
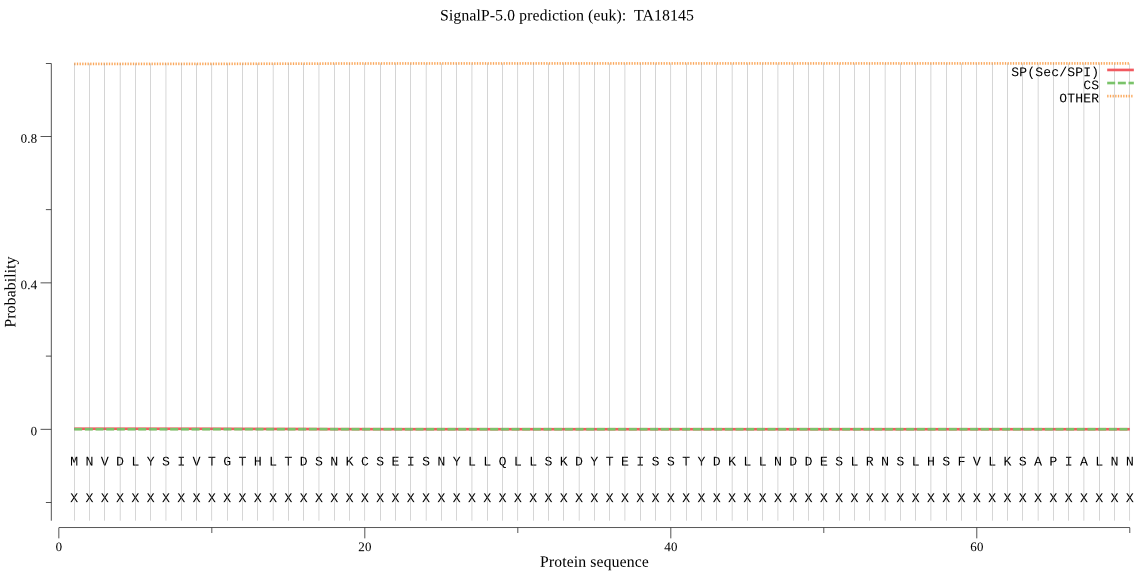
Fasta :-
>TA18145
MNVDLYSIVTGTHLTDSNKCSEISNYLLQLLSKDYTEISSTYDKLLNDDESLRNSLHSFV
LKSAPIALNNINYTNNSLYHVGNLSKIAFDIKSSNLLSHKSTELEESLTRIVEELTEINS
EYPDYKYLNSLLNFLNNFELNDTNLSFLLSINKVLNSNDSIQSELGLYLSNKVRNLLVSV
EFELFTSFSNDFDENTKIIELIKKIGLSSVHEISVGFLMNRFDYLNNSFNSCVLKSNVNI
QRNVFGDIDVNLSTFVNRVFQEFFDTIIKINSQDKLELDLNLSQLDEELINLSKLLSPLG
LSFHHITNFTLTNIINI
- Download Fasta
-
MitoProt II - v1.101
File : /home/rajan/sadaf/4480_mitoprot/test/TA18145.fa
Sequence name : TA18145
Sequence length : 317
VALUES OF COMPUTED PARAMETERS
Coef20 : 3.724
CoefTot : -0.134
ChDiff : -17
ZoneTo : 3
KR : 0
DE : 0
CleavSite : 0
HYDROPHOBIC SCALE USED
GES KD GVH1 ECS
H17 : 1.065 0.859 -0.125 0.462
MesoH : -0.900 0.270 -0.504 0.175
MuHd_075 : 9.256 12.246 3.247 2.641
MuHd_095 : 4.487 3.589 1.972 0.503
MuHd_100 : 4.222 4.068 2.124 0.852
MuHd_105 : 6.914 5.179 2.118 1.425
Hmax_075 : 16.333 14.350 2.260 4.480
Hmax_095 : 4.600 6.700 -0.736 2.390
Hmax_100 : 5.100 9.500 -0.018 3.240
Hmax_105 : 9.500 9.500 0.297 3.360
CLASS NOT-MITO MITO(/CHLORO)
DFM : 0.9549 0.0451
DFMC : 0.8859 0.1141
-
Fasta :-
>TA18145
ATGAATGTTGATTTGTATTCTATAGTTACTGGTACTCATTTAACAGATTCTAACAAGTGT
AGTGAAATTTCCAATTACCTTCTACAATTATTATCCAAAGATTATACTGAAATTTCATCT
ACATATGATAAATTATTAAATGATGATGAATCACTACGGAATAGTTTACACAGTTTCGTT
CTAAAATCTGCTCCTATCGCTTTAAATAATATTAATTATACCAACAATTCATTATATCAT
GTTGGGAATTTATCAAAAATTGCTTTTGATATTAAATCTTCTAATTTATTATCACATAAA
TCTACGGAGTTGGAAGAATCATTAACTAGAATTGTTGAAGAATTAACAGAGATTAATTCG
GAATATCCTGATTACAAATATTTAAATTCATTATTAAACTTTTTAAACAATTTTGAATTG
AATGATACAAATTTGTCATTTTTACTATCAATCAATAAGGTTCTAAACTCTAATGATTCT
ATTCAAAGTGAATTAGGTTTATATCTATCAAACAAAGTTAGAAACTTGTTAGTTTCAGTG
GAATTTGAGTTATTTACATCATTTAGTAATGATTTTGATGAGAATACTAAGATAATAGAA
CTTATTAAGAAAATTGGTTTATCAAGTGTTCATGAAATATCTGTTGGTTTTCTTATGAAT
AGATTTGATTATTTGAATAATTCCTTCAATTCCTGTGTACTCAAATCAAATGTTAATATT
CAAAGAAATGTGTTTGGTGATATTGATGTAAATTTATCAACATTTGTGAACAGAGTATTT
CAAGAGTTTTTTGATACAATAATAAAAATAAATTCACAAGATAAATTGGAACTGGATTTA
AATTTATCACAACTTGATGAAGAATTAATAAATTTATCAAAATTATTATCACCATTAGGA
TTATCATTTCATCATATTACTAATTTTACATTAACCAATATTATTAATATATAA
- Download Fasta
|
ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method |
|
TA18145 | 17 S | HLTDSNKCS | 0.992 | unsp | TA18145 | 98 S | SNLLSHKST | 0.994 | unsp |


