

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| TA18155 | SP | 0.435028 | 0.492663 | 0.072309 | CS pos: 26-27. CVT-IQ. Pr: 0.4948 |
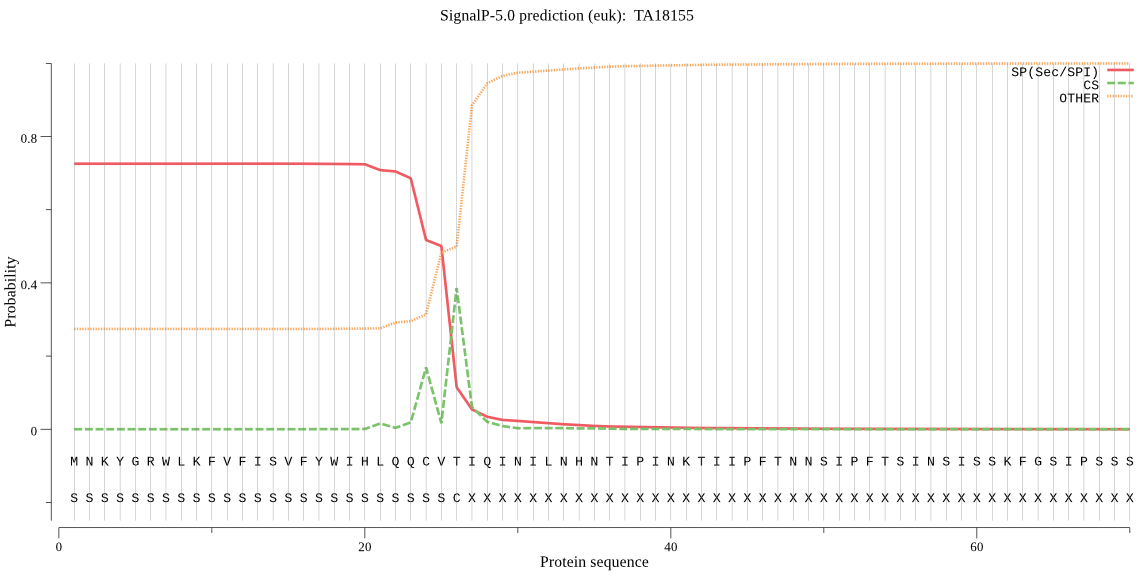
MNKYGRWLKFVFISVFYWIHLQQCVTIQINILNHNTIPINKTIIPFTNNSIPFTSINSIS SKFGSIPSSSINSVSSKFGSVPSSSINSIPFTNINHVPYKFGRLGFINTNVLKFTKFNTN YNLTECKLVKDKSITHLDKQQTPLPNELKKQSLTTNLSLYVSKFLKNVKESNPYKYLTVK LSKFCVMIFGSALHTLVLSKCRFVLPYQLIPFNSGLGTEVTLDTLISIPILYYILKDYTF TQSTLETNNLKPKLEDNNLKSKLEDNKLTNKSKMVNINQTSLDEKYFQLPVRSKFKISMV VTSLLGSFIVSSYFSQFIDSLLLLVSAFDFPISVGMLKSFSVMSSHLFWVFIGSLILKSC MFPIFSKSNPWYKLDLSKLWVHQVLMGYYSSYTIYKLADIGYYLFLKFFDIQNQVSLTDE QISNICETNETIPILITSIGPCLTAPWWEELLYRVFTFKLFNSFLPTLYSSIASSIIFSI NHLSPHSFLQLFSLGLLWSFIQLNNDNILITILIHSLWNSRIFFGSLTGI