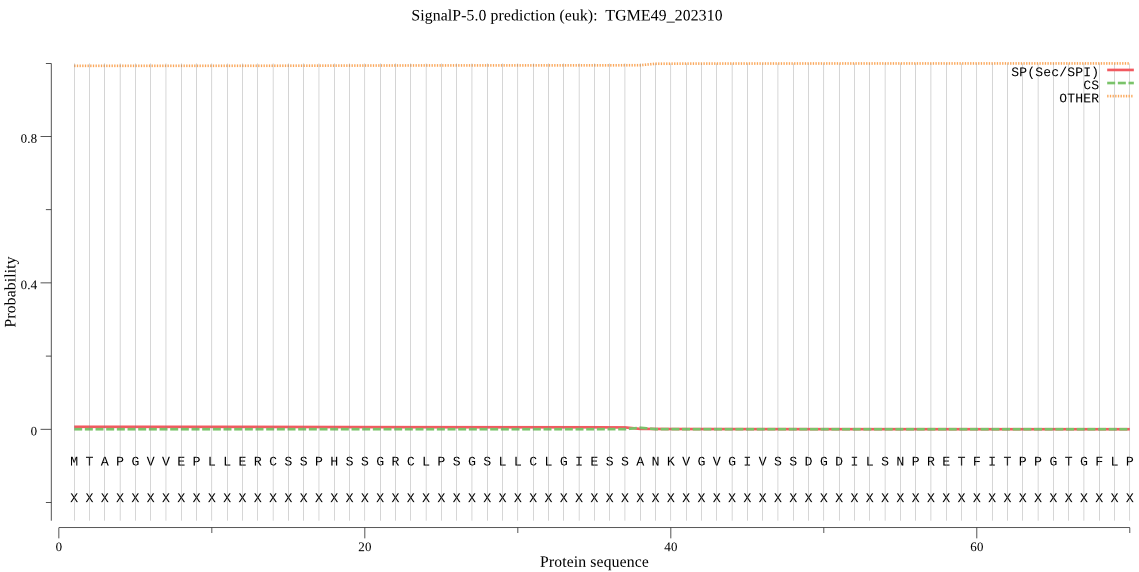
Fasta :-
MTAPGVVEPLLERCSSPHSSGRCLPSGSLLCLGIESSANKVGVGIVSSDGDILSNPRETF
ITPPGTGFLPRETAAHHQGKIVGLVRRALTEARVEPKQLSCIAYTCGPGMGGPLAVGAIT
ARTLSLLWNIPLVAVNHCVAHIEMGRLVTGCANPVVLYVSGGNTQVIGYADGRYRILGET
LDVAVGNCIDRLARLLHLPNDPAPGYQVEQLARRFLETKRKRSSFTDSLKTPGGGSQIEE
PAQGRIERTQEDHTEMLLPLPYTVKGMDLSFSGILTRLEDIAGTMRRYEKFRNEMRQDCE
PEVDCILSSKHAKQESRGPALVGTHEPKQSHELSHYGEQKSQAGCCSVEADTGGPTGRPP
PSADANAGTAGTSDDCFEGPASSCKQRGSDGKAAEKQQRRRGCSGRKEESELRAYPQGLQ
GIQGMKKLREGSCAAKGNSKRQCQSVDMFEGLPTCLLTPESLCFSAQEIIFAMLTEVTER
AMALHYADQVLVVGGVGCNLRLQEMLKEMAMRRGASMGGMDDRYCIDNGAMVAYLGCLMA
SKGQFVDVSKAHYRQRFRTDEVPVLWRENDNSHELAEMQG