

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| TGME49_202680 | mTP | 0.045073 | 0.001018 | 0.953909 | CS pos: 55-56. KRF-LS. Pr: 0.1124 |
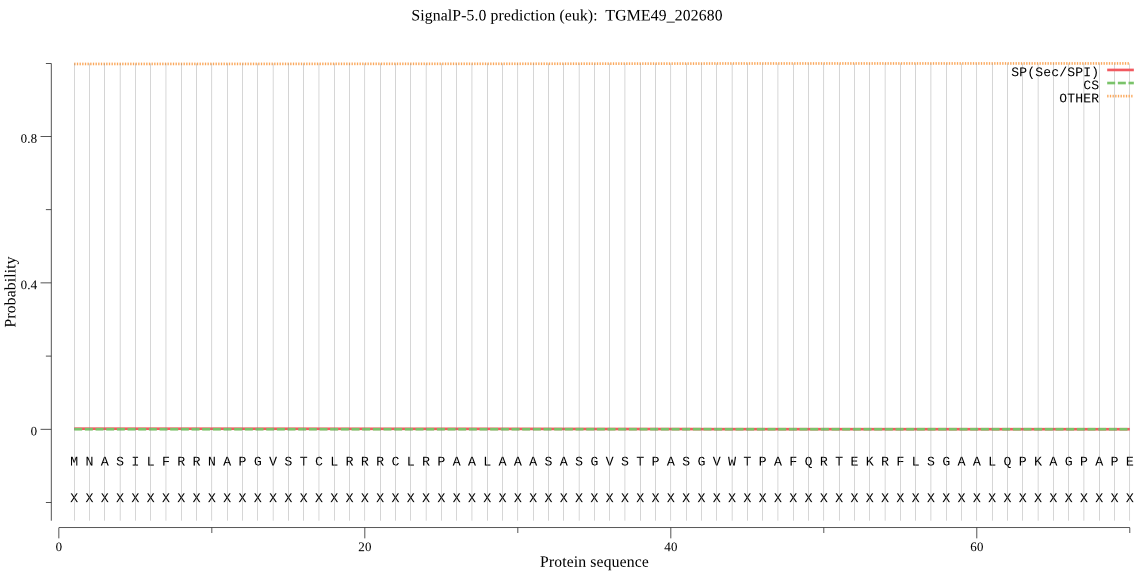
MNASILFRRNAPGVSTCLRRRCLRPAALAAASASGVSTPASGVWTPAFQRTEKRFLSGAA LQPKAGPAPEYRRVPFVKEDMEKVMEEVPEFKYYYVGKENTKGNVYEGIPLDQSILEPAD LRDYVPPHSNIQYSKLDNGLRIASMDRGGLTASLGLFVHAGTRFEDVTNFGVTHMIQNLA FASTAHLSLLRTVKTIEVLGANAGCVVGREHLVYSAECLRSHMPLLVPMLTGNVLFPRFL PWELKACKEKLIMARKRLEHMPDQMVSELLHTTAWHNNTLGHKLHCTERSLGHYNPDVIR HYMLQHFSPENMVFVGVNVNHDELCTWLMRAFVDYNAIPPSKRTVASPVYTGGDVRLETP SPHAHMAIAFETPGGWNGGDLVAYSVLQTILGGGGAFSTGGPGKGMYTRLYLNVLNQNEW VESAMAFNTQYTDSGIFGLYMLADPTKSANAVKVMAEQFGKMVSVTKEELQRAKNSLKSS IFMNLECRGIVMEDVGRQLLMSNRVISPQEFCTAIDAVTEADIKRVVDAMYKKPPTVVAY GDVSTVPHYEEVRAALRAAGVGK
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| TGME49_202680 | 507 S | NRVISPQEF | 0.996 | unsp |