

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
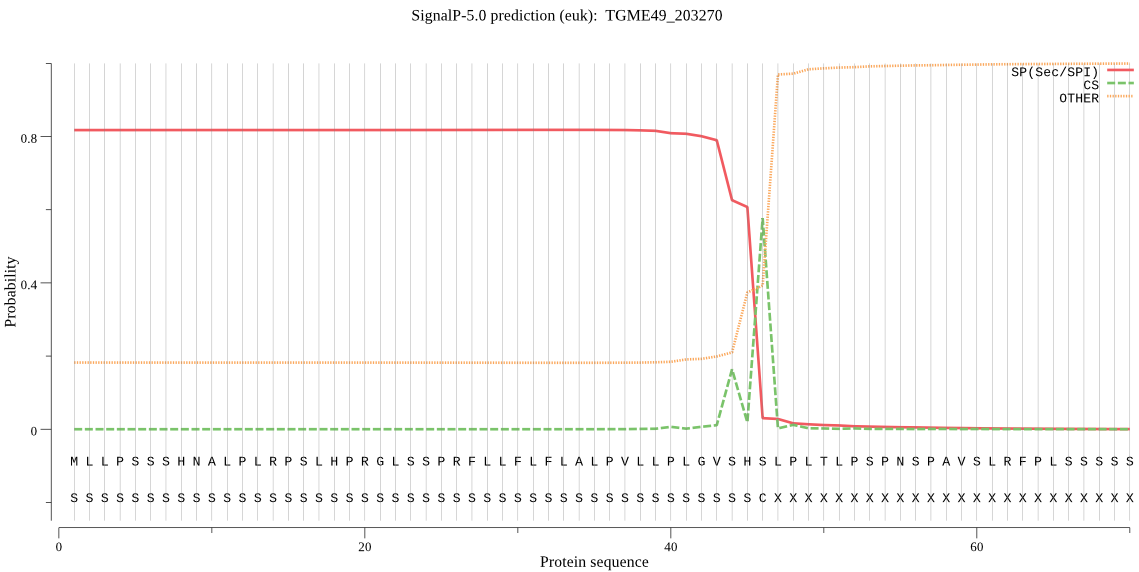
| TGME49_203270 | SP | 0.293931 | 0.705790 | 0.000280 | CS pos: 46-47. SHS-LP. Pr: 0.5455 |
MLLPSSSHNALPLRPSLHPRGLSSPRFLLFLFLALPVLLPLGVSHSLPLTLPSPNSPAVS LRFPLSSSSSLASSSASPLFSFSRLSRLRSSLSPHPCSLTPVSSCSRAAADVDGLSFCPV SVAPLTPRHSVSLAVSPLCRSRKRDVRGVEPRFLPRFLLLSGASASRRARLSCRDANAQS GLASALPSSLWGPLFVATIAAVPCSLRCLNIAKKSSTRKSRDLCFARHAGISLRKRITEK TTPLLYGATVARDKSRRNASSSFISSPLSSICHLCSFPSSSLSPVSAFSSSSSSSFSPLS SSFASTPMSFFLKRNCNACWGREKDAGNRLSLGETEALADSSETRRRSTLAFALRHSFLC PSLTRSSSRASLPFQASSSSSHSRLSGALSSPHQLSERRPLASWRCLYSLLEREARRRQE GGARVFFLSQDLTEDLAHRLIAHLFFHDRGNRKKCNVRSGCESSQPCQALPEASLASLPS SSSSSSLPPSSSASPASTASSSFSASPCTSSSLSALSSSACSSASSSCFVSPPASREEAR QRDVTCLLRRLTATRGEDSRLAPSADRRLWNRSTGGGDSDVDVDRPRKTGGNQEEGDERT ADADPFVIFVNSAGGSLSAGLALFDALSTLQAPVYTVNLGLAASAASLLLAAGSGGRRFA VEQSSVLLHQPAGSLAGRDSELQDERREVEKLHEKVVRLYRCETAEEATRSERTERDTNV LTRKREESRREREEDDRPSDGHNDEVTGRPEARVRRDIKEEKVLTAEQACEYGLVDAVLP LFDAKEIRA
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TGME49_203270 | 216 S | AKKSSTRKS | 0.995 | unsp | TGME49_203270 | 216 S | AKKSSTRKS | 0.995 | unsp | TGME49_203270 | 216 S | AKKSSTRKS | 0.995 | unsp | TGME49_203270 | 255 S | ARDKSRRNA | 0.994 | unsp | TGME49_203270 | 283 S | SSSLSPVSA | 0.992 | unsp | TGME49_203270 | 331 S | GNRLSLGET | 0.996 | unsp | TGME49_203270 | 348 S | TRRRSTLAF | 0.993 | unsp | TGME49_203270 | 367 S | LTRSSSRAS | 0.994 | unsp | TGME49_203270 | 381 S | SSSSSHSRL | 0.992 | unsp | TGME49_203270 | 535 S | SPPASREEA | 0.997 | unsp | TGME49_203270 | 579 S | GGGDSDVDV | 0.99 | unsp | TGME49_203270 | 714 T | RSERTERDT | 0.992 | unsp | TGME49_203270 | 728 S | KREESRRER | 0.998 | unsp | TGME49_203270 | 739 S | DDRPSDGHN | 0.994 | unsp | TGME49_203270 | 91 S | RLRSSLSPH | 0.993 | unsp | TGME49_203270 | 172 S | RARLSCRDA | 0.998 | unsp |