

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| TGME49_203300 | OTHER | 0.999946 | 0.000050 | 0.000004 |
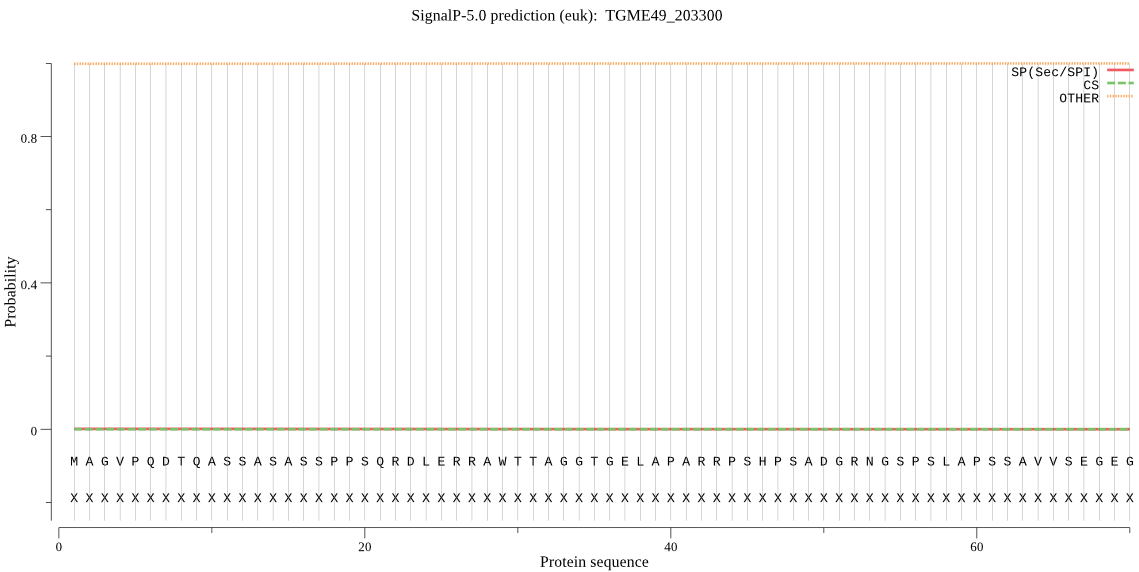
MAGVPQDTQASSASASSPPSQRDLERRAWTTAGGTGELAPARRPSHPSADGRNGSPSLAP SSAVVSEGEGGMTHSVVSSSTMPFGRFSSLASSCQDNADEPICFLQPDQQQHFLLQGRLV DDTISRPARSLFSLVSGAASDDAPAPHHAAAAASAGVFVHPSGALQSAEPGSTANRAAEA AGSPCLRQGSSLFSSASGASGRSLGLSRSVEKIIESGEVQMDDIEPEQAADISKALLLHT QHHHMLLQSESLPRTPLTVHPLSPFRQKEPGAERGTPTAGGFASAERGTRTLNIIREVYE GYQSSSPSSARHLSEDGRGGEDGRGGEDGRGGEDGRGGEDGRGGEDGRGGEAEALMKASP RDEAPETAETRRSTHEREKDKAVSVHARPGTEDEGSGASGQLSAPREYRQAASDAEKGHS GAHTSSRQDREDRGGGVALPSETEEEQERRLLDEIEARLTQDAEGFLEGAKLELEDLRLS AEREVLALQYIPFKADRLLALPTSCRESLLGIFVCLCLQHIESLSPELQSQVAEWSLIFL HTLATPQVLNVDANVEKSLVRPIILRDRLLKDGLKTSDILSPAPLVKVVADACEAMQPEV NSIFNTLHSEMSCFAHLGISQDSAPASPVAPGAAGAQRPRTRSQSPSFSSSEAADSSAGV EPRRRSSRRDILSSLPAAPHAQYARERQGIMDDSEEDGARPSGAQRVEGVEPRGVAAYVY SSGLRRGSDASDAERGATIGLGRRGEEEDSSFLEEAGQTVSEHLRRRLTVERRREELHAR RREAAERAEDGGEETMERSRRRENDEELLSSPRENGEPGAPPDTASVDRNSESQTLASSS VAASSPSVGCRHDSHAVVFTSVAAHPLSGGSSPLSGSARTEAPHAAATLRVCVEKREEED EALVAAVGCVEEKESFDEEGSGCELQLPEEQEQMLNPNSLRSEQPTRYNFPAFSRAQSGR TSWNPSPWERRSPEGTFGSEDGASALLPSYPPSAKTVLLRELTTALVTSGSFDARVQFML QVFAVDLLHLNPLILIRLEHHLAADLLDLLRASTKEGARQKTWRRLKIAGAAIGGGVLVA LTAGLAAPGIVAGIASLGLGGAGVSAFLASAGGMAVIVSLFGAGGAGLTGWKYSRRIANI KIFEFDMLNGKTPSSLGVTVCVSGYLRTFDDIALPWLQGCPHACSDLLALKWEPNILKSL GGMVVQMLSQDFAVAAGKYYLQHTALGGLTFALFWPMALIQYAASLDNTWSLCRERAQQA GRLLADAISDTQAVGQRPVSLVGYSMGARVIFYCLQTLWKRRKLHVVCDCVLIGLPASLN TKQWSAARDVVSRRLINVYSRSDWLLAFLYRWMEWGLQVAGLGPVKNVPGVENYDVTGLV KSHAQYPEMMPDILAFIGFDA
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TGME49_203300 | 45 S | ARRPSHPSA | 0.997 | unsp | TGME49_203300 | 45 S | ARRPSHPSA | 0.997 | unsp | TGME49_203300 | 45 S | ARRPSHPSA | 0.997 | unsp | TGME49_203300 | 263 S | VHPLSPFRQ | 0.996 | unsp | TGME49_203300 | 306 S | YQSSSPSSA | 0.993 | unsp | TGME49_203300 | 359 S | LMKASPRDE | 0.996 | unsp | TGME49_203300 | 373 S | ETRRSTHER | 0.997 | unsp | TGME49_203300 | 480 S | DLRLSAERE | 0.997 | unsp | TGME49_203300 | 643 S | PRTRSQSPS | 0.993 | unsp | TGME49_203300 | 645 S | TRSQSPSFS | 0.997 | unsp | TGME49_203300 | 666 S | PRRRSSRRD | 0.998 | unsp | TGME49_203300 | 667 S | RRRSSRRDI | 0.998 | unsp | TGME49_203300 | 694 S | IMDDSEEDG | 0.997 | unsp | TGME49_203300 | 811 S | ELLSSPREN | 0.996 | unsp | TGME49_203300 | 831 S | VDRNSESQT | 0.995 | unsp | TGME49_203300 | 962 S | SGRTSWNPS | 0.99 | unsp | TGME49_203300 | 972 S | WERRSPEGT | 0.998 | unsp | TGME49_203300 | 1053 S | LLRASTKEG | 0.998 | unsp | TGME49_203300 | 17 S | ASASSPPSQ | 0.993 | unsp | TGME49_203300 | 20 S | SSPPSQRDL | 0.998 | unsp |